Lymphocyte-monocyte-neutrophil index: a predictor of severity of coronavirus disease 2019 patients produced by sparse principal component analysis
- PMID: 34088324
- PMCID: PMC8176446
- DOI: 10.1186/s12985-021-01561-9
Lymphocyte-monocyte-neutrophil index: a predictor of severity of coronavirus disease 2019 patients produced by sparse principal component analysis
Abstract
Background: It is important to recognize the coronavirus disease 2019 (COVID-19) patients in severe conditions from moderate ones, thus more effective predictors should be developed.
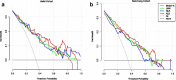
Methods: Clinical indicators of COVID-19 patients from two independent cohorts (Training data: Hefei Cohort, 82 patients; Validation data: Nanchang Cohort, 169 patients) were retrospected. Sparse principal component analysis (SPCA) using Hefei Cohort was performed and prediction models were deduced. Prediction results were evaluated by receiver operator characteristic curve and decision curve analysis (DCA) in above two cohorts.
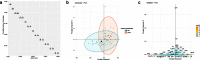
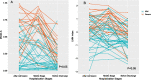
Results: SPCA using Hefei Cohort revealed that the first 13 principal components (PCs) account for 80.8% of the total variance of original data. The PC1 and PC12 were significantly associated with disease severity with odds ratio of 4.049 and 3.318, respectively. They were used to construct prediction model, named Model-A. In disease severity prediction, Model-A gave the best prediction efficiency with area under curve (AUC) of 0.867 and 0.835 in Hefei and Nanchang Cohort, respectively. Model-A's simplified version, named as LMN index, gave comparable prediction efficiency as classical clinical markers with AUC of 0.837 and 0.800 in training and validation cohort, respectively. According to DCA, Model-A gave slightly better performance than others and LMN index showed similar performance as albumin or neutrophil-to-lymphocyte ratio.
Conclusions: Prediction models produced by SPCA showed robust disease severity prediction efficiency for COVID-19 patients and have the potential for clinical application.
Keywords: COVID-19; Prediction; Principal component analysis; SARS-CoV-2; Severity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
A meta-analysis of SARS-CoV-2 patients identifies the combinatorial significance of D-dimer, C-reactive protein, lymphocyte, and neutrophil values as a predictor of disease severity.Int J Lab Hematol. 2021 Apr;43(2):324-328. doi: 10.1111/ijlh.13354. Epub 2020 Oct 3. Int J Lab Hematol. 2021. PMID: 33010111 Free PMC article.
-
Diagnostic value of peripheral hematologic markers for coronavirus disease 2019 (COVID-19): A multicenter, cross-sectional study.J Clin Lab Anal. 2020 Oct;34(10):e23475. doi: 10.1002/jcla.23475. Epub 2020 Jul 17. J Clin Lab Anal. 2020. PMID: 32681559 Free PMC article.
-
Evaluation of simple and cost-effective immuno- haematological markers to predict outcome in hospitalized severe COVID-19 patients, with a focus on diabetes mellitus - A retrospective study in Andhra Pradesh, India.Diabetes Metab Syndr. 2021 May-Jun;15(3):739-745. doi: 10.1016/j.dsx.2021.03.025. Epub 2021 Mar 29. Diabetes Metab Syndr. 2021. PMID: 33819728 Free PMC article.
-
Prediction of the risk of mortality in older patients with coronavirus disease 2019 using blood markers and machine learning.Front Immunol. 2024 Nov 1;15:1445618. doi: 10.3389/fimmu.2024.1445618. eCollection 2024. Front Immunol. 2024. PMID: 39555074 Free PMC article.
-
Immunity and coagulation and fibrinolytic processes may reduce the risk of severe illness in pregnant women with coronavirus disease 2019.Am J Obstet Gynecol. 2021 Apr;224(4):393.e1-393.e25. doi: 10.1016/j.ajog.2020.10.032. Epub 2020 Oct 22. Am J Obstet Gynecol. 2021. PMID: 33098813 Free PMC article.
Cited by
-
Selenium Antagonizes Cadmium-Induced Inflammation and Oxidative Stress via Suppressing the Interplay between NLRP3 Inflammasome and HMGB1/NF-κB Pathway in Duck Hepatocytes.Int J Mol Sci. 2022 Jun 2;23(11):6252. doi: 10.3390/ijms23116252. Int J Mol Sci. 2022. PMID: 35682929 Free PMC article.
-
Application of Principal Component Analysis to Heterogenous Fontan Registry Data Identifies Independent Contributing Factors to Decline.medRxiv [Preprint]. 2024 Jul 12:2024.07.11.24310309. doi: 10.1101/2024.07.11.24310309. medRxiv. 2024. PMID: 39040194 Free PMC article. Preprint.
-
Value of the Neutrophil-Lymphocyte Ratio in Predicting COVID-19 Severity: A Meta-analysis.Dis Markers. 2021 Oct 8;2021:2571912. doi: 10.1155/2021/2571912. eCollection 2021. Dis Markers. 2021. PMID: 34650648 Free PMC article.
-
Laboratory biomarkers associated with COVID-19 mortality among inpatients in a Peruvian referral hospital.Heliyon. 2024 Feb 29;10(6):e27251. doi: 10.1016/j.heliyon.2024.e27251. eCollection 2024 Mar 30. Heliyon. 2024. PMID: 38500972 Free PMC article.
-
Neutrophil count predicts clinical outcome in hospitalized COVID-19 patients: Results from the NOR-Solidarity trial.J Intern Med. 2022 Feb;291(2):241-243. doi: 10.1111/joim.13377. Epub 2021 Oct 3. J Intern Med. 2022. PMID: 34411368 Free PMC article. No abstract available.
References
-
- WHO. Coronavirus disease 2019 (COVID-19) situation report-130. https://www.who.int/docs/default-source/coronaviruse/situation-reports/2.... Accessed 29 May 2020.
-
- Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. 10.3760/cma.j.issn.0254-6450.2020.02.003. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

