Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation
- PMID: 34088835
- PMCID: PMC8201911
- DOI: 10.1073/pnas.2022704118
Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation
Abstract
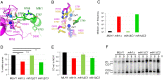
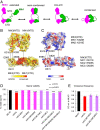
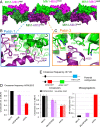
In budding yeast, the MutL homolog heterodimer Mlh1-Mlh3 (MutLγ) plays a central role in the formation of meiotic crossovers. It is also involved in the repair of a subset of mismatches besides the main mismatch repair (MMR) endonuclease Mlh1-Pms1 (MutLα). The heterodimer interface and endonuclease sites of MutLγ and MutLα are located in their C-terminal domain (CTD). The molecular basis of MutLγ's dual roles in MMR and meiosis is not known. To better understand the specificity of MutLγ, we characterized the crystal structure of Saccharomyces cerevisiae MutLγ(CTD). Although MutLγ(CTD) presents overall similarities with MutLα(CTD), it harbors some rearrangement of the surface surrounding the active site, which indicates altered substrate preference. The last amino acids of Mlh1 participate in the Mlh3 endonuclease site as previously reported for Pms1. We characterized mlh1 alleles and showed a critical role of this Mlh1 extreme C terminus both in MMR and in meiotic recombination. We showed that the MutLγ(CTD) preferentially binds Holliday junctions, contrary to MutLα(CTD). We characterized Mlh3 positions on the N-terminal domain (NTD) and CTD that could contribute to the positioning of the NTD close to the CTD in the context of the full-length MutLγ. Finally, crystal packing revealed an assembly of MutLγ(CTD) molecules in filament structures. Mutation at the corresponding interfaces reduced crossover formation, suggesting that these superstructures may contribute to the oligomer formation proposed for MutLγ. This study defines clear divergent features between the MutL homologs and identifies, at the molecular level, their specialization toward MMR or meiotic recombination functions.
Keywords: DNA recombination; DNA repair; biochemistry; genetics; structural biology.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Distinct DNA-binding surfaces in the ATPase and linker domains of MutLγ determine its substrate specificities and exert separable functions in meiotic recombination and mismatch repair.PLoS Genet. 2017 May 15;13(5):e1006722. doi: 10.1371/journal.pgen.1006722. eCollection 2017 May. PLoS Genet. 2017. PMID: 28505149 Free PMC article.
-
The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans.PLoS Biol. 2017 Apr 28;15(4):e2001164. doi: 10.1371/journal.pbio.2001164. eCollection 2017 Apr. PLoS Biol. 2017. PMID: 28453523 Free PMC article.
-
The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions.J Biol Chem. 2014 Feb 28;289(9):5674-86. doi: 10.1074/jbc.M113.533810. Epub 2014 Jan 17. J Biol Chem. 2014. PMID: 24443562 Free PMC article.
-
Expanded roles for the MutL family of DNA mismatch repair proteins.Yeast. 2021 Jan;38(1):39-53. doi: 10.1002/yea.3512. Epub 2020 Jul 30. Yeast. 2021. PMID: 32652606 Free PMC article. Review.
-
Coordinated and Independent Roles for MLH Subunits in DNA Repair.Cells. 2021 Apr 20;10(4):948. doi: 10.3390/cells10040948. Cells. 2021. PMID: 33923939 Free PMC article. Review.
Cited by
-
The Dmc1 recombinase physically interacts with and promotes the meiotic crossover functions of the Mlh1-Mlh3 endonuclease.Genetics. 2024 Jul 8;227(3):iyae066. doi: 10.1093/genetics/iyae066. Genetics. 2024. PMID: 38657110 Free PMC article.
-
Untangle the knot: Soybean MLH1 in meiotic recombination.Plant Physiol. 2024 Sep 2;196(1):30-31. doi: 10.1093/plphys/kiae266. Plant Physiol. 2024. PMID: 38718098 Free PMC article. No abstract available.
-
Integration of deep learning with Ramachandran plot molecular dynamics simulation for genetic variant classification.iScience. 2023 Feb 2;26(3):106122. doi: 10.1016/j.isci.2023.106122. eCollection 2023 Mar 17. iScience. 2023. PMID: 36879825 Free PMC article.
-
Catalytic mechanism of the zinc-dependent MutL endonuclease reaction.Life Sci Alliance. 2023 Jul 24;6(10):e202302001. doi: 10.26508/lsa.202302001. Print 2023 Oct. Life Sci Alliance. 2023. PMID: 37487639 Free PMC article.
-
Therapeutic validation of MMR-associated genetic modifiers in a human ex vivo model of Huntington disease.Am J Hum Genet. 2024 Jun 6;111(6):1165-1183. doi: 10.1016/j.ajhg.2024.04.015. Epub 2024 May 14. Am J Hum Genet. 2024. PMID: 38749429 Free PMC article.
References
-
- Allers T., Lichten M., Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001). - PubMed
-
- Hunter N., Kleckner N., The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

