Sequence of the supernumerary B chromosome of maize provides insight into its drive mechanism and evolution
- PMID: 34088847
- PMCID: PMC8201846
- DOI: 10.1073/pnas.2104254118
Sequence of the supernumerary B chromosome of maize provides insight into its drive mechanism and evolution
Abstract
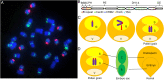
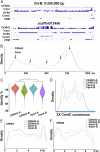
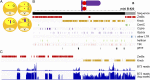
B chromosomes are enigmatic elements in thousands of plant and animal genomes that persist in populations despite being nonessential. They circumvent the laws of Mendelian inheritance but the molecular mechanisms underlying this behavior remain unknown. Here we present the sequence, annotation, and analysis of the maize B chromosome providing insight into its drive mechanism. The sequence assembly reveals detailed locations of the elements involved with the cis and trans functions of its drive mechanism, consisting of nondisjunction at the second pollen mitosis and preferential fertilization of the egg by the B-containing sperm. We identified 758 protein-coding genes in 125.9 Mb of B chromosome sequence, of which at least 88 are expressed. Our results demonstrate that transposable elements in the B chromosome are shared with the standard A chromosome set but multiple lines of evidence fail to detect a syntenic genic region in the A chromosomes, suggesting a distant origin. The current gene content is a result of continuous transfer from the A chromosomal complement over an extended evolutionary time with subsequent degradation but with selection for maintenance of this nonvital chromosome.
Keywords: B chromosome; genetic drive; nondisjunction; preferential fertilization.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: G.B.-Z., L.G., G.K., and K.B. are employees of NRGene. J.C.L. is an employee of Bayer Crop Science.
Figures


References
-
- Wilson E. B., The supernumerary chromosomes of Hemiptera. Science 26, 870–871 (1907).
-
- D’Ambrosio U., et al. ., B-chrom: A database on B-chromosomes of plants, animals and fungi. New Phytol. 216, 635–642 (2017). - PubMed
-
- Werren J. H., Nur U., Eickbush D., An extrachromosomal factor causing loss of paternal chromosomes. Nature 327, 75–76 (1987). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

