Homologous and heterologous serological response to the N-terminal domain of SARS-CoV-2 in humans and mice
- PMID: 34089541
- PMCID: PMC8237060
- DOI: 10.1002/eji.202149234
Homologous and heterologous serological response to the N-terminal domain of SARS-CoV-2 in humans and mice
Abstract
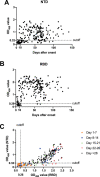
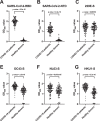
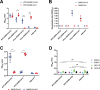
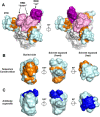
The increasing numbers of infected cases of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses serious threats to public health and the global economy. Most SARS-CoV-2 neutralizing antibodies target the receptor binding domain (RBD) and some the N-terminal domain (NTD) of the spike protein, which is the major antigen of SARS-CoV-2. While the antibody response to RBD has been extensively characterized, the antigenicity and immunogenicity of the NTD protein are less well studied. Using 227 plasma samples from COVID-19 patients, we showed that SARS-CoV-2 NTD-specific antibodies could be induced during infection. As compared to the results of SARS-CoV-2 RBD, the serological response of SARS-CoV-2 NTD is less cross-reactive with SARS-CoV, a pandemic strain that was identified in 2003. Furthermore, neutralizing antibodies are rarely elicited in a mice model when NTD is used as an immunogen. We subsequently demonstrate that NTD has an altered antigenicity when expressed alone. Overall, our results suggest that while NTD offers a supplementary strategy for serology testing, it may not be suitable as an immunogen for vaccine development.
Keywords: COVID-19; N-terminal domain; NTD; SARS-CoV-2; immunogen; serology.
© 2021 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no financial or commercial conflict of interest.
Figures

Similar articles
-
Neutralizing antibody response elicited by SARS-CoV-2 receptor-binding domain.Hum Vaccin Immunother. 2021 Mar 4;17(3):654-655. doi: 10.1080/21645515.2020.1814098. Epub 2020 Sep 29. Hum Vaccin Immunother. 2021. PMID: 32991231 Free PMC article.
-
Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections.Cell Rep. 2020 Jun 2;31(9):107725. doi: 10.1016/j.celrep.2020.107725. Epub 2020 May 18. Cell Rep. 2020. PMID: 33500101
-
A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.Cell. 2020 Aug 6;182(3):722-733.e11. doi: 10.1016/j.cell.2020.06.035. Epub 2020 Jun 28. Cell. 2020. PMID: 32645327 Free PMC article.
-
An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.Cell. 2021 Jun 24;184(13):3452-3466.e18. doi: 10.1016/j.cell.2021.05.032. Epub 2021 May 24. Cell. 2021. PMID: 34139176 Free PMC article.
-
Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies.Biochem Biophys Res Commun. 2021 Jan 29;538:192-203. doi: 10.1016/j.bbrc.2020.10.012. Epub 2020 Oct 10. Biochem Biophys Res Commun. 2021. PMID: 33069360 Free PMC article. Review.
Cited by
-
Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.Cell Host Microbe. 2023 Jan 11;31(1):97-111.e12. doi: 10.1016/j.chom.2022.10.010. Epub 2022 Nov 7. Cell Host Microbe. 2023. PMID: 36347257 Free PMC article.
-
Antibody-mediated immunity to SARS-CoV-2 spike.Adv Immunol. 2022;154:1-69. doi: 10.1016/bs.ai.2022.07.001. Epub 2022 Aug 22. Adv Immunol. 2022. PMID: 36038194 Free PMC article. Review.
-
Broadly neutralizing antibodies target the coronavirus fusion peptide.bioRxiv [Preprint]. 2022 Apr 12:2022.04.11.487879. doi: 10.1101/2022.04.11.487879. bioRxiv. 2022. Update in: Science. 2022 Aug 12;377(6607):728-735. doi: 10.1126/science.abq3773. PMID: 35441178 Free PMC article. Updated. Preprint.
-
Structural Immunology of SARS-CoV-2.Immunol Rev. 2025 Jan;329(1):e13431. doi: 10.1111/imr.13431. Epub 2024 Dec 27. Immunol Rev. 2025. PMID: 39731211 Free PMC article. Review.
References
-
- Pinto, D. , Park, Y. J. , Beltramello, M. , Walls, A. C. , Tortorici, M. A. , Bianchi, S. , Jaconi, S. et al., Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature 2020. 583: 290–295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

