Phosphorylation of human phospholipase A1 DDHD1 at newly identified phosphosites affects its subcellular localization
- PMID: 34089703
- PMCID: PMC8234217
- DOI: 10.1016/j.jbc.2021.100851
Phosphorylation of human phospholipase A1 DDHD1 at newly identified phosphosites affects its subcellular localization
Abstract
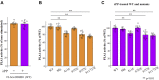
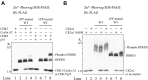
Phospholipase A1 (PLA1) hydrolyzes the fatty acids of glycerophospholipids, which are structural components of the cellular membrane. Genetic mutations in DDHD1, an intracellular PLA1, result in hereditary spastic paraplegia (HSP) in humans. However, the regulation of DDHD1 activity has not yet been elucidated in detail. In the present study, we examined the phosphorylation of DDHD1 and identified the responsible protein kinases. We performed MALDI-TOF MS/MS analysis and Phos-tag SDS-PAGE in alanine-substitution mutants in HEK293 cells and revealed multiple phosphorylation sites in human DDHD1, primarily Ser8, Ser11, Ser723, and Ser727. The treatment of cells with a protein phosphatase inhibitor induced the hyperphosphorylation of DDHD1, suggesting that multisite phosphorylation occurred not only at these major, but also at minor sites. Site-specific kinase-substrate prediction algorithms and in vitro kinase analyses indicated that cyclin-dependent kinase CDK1/cyclin A2 phosphorylated Ser8, Ser11, and Ser727 in DDHD1 with a preference for Ser11 and that CDK5/p35 also phosphorylated Ser11 and Ser727 with a preference for Ser11. In addition, casein kinase CK2α1 was found to phosphorylate Ser104, although this was not a major phosphorylation site in cultivated HEK293 cells. The evaluation of the effects of phosphorylation revealed that the phosphorylation mimic mutants S11/727E exhibit only 20% reduction in PLA1 activity. However, the phosphorylation mimics were mainly localized to focal adhesions, whereas the phosphorylation-resistant mutants S11/727A were not. This suggested that phosphorylation alters the subcellular localization of DDHD1 without greatly affecting its PLA1 activity.
Keywords: DDHD domain containing 1 (DDHD1); Phos-tag; cyclin-dependent kinase (CDK); hereditary spastic paraplegia (HSP); phospholipase A1 (PLA1); phosphorylation.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest regarding the content of this article.
Figures






Similar articles
-
Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles.Molecules. 2022 Apr 12;27(8):2487. doi: 10.3390/molecules27082487. Molecules. 2022. PMID: 35458682 Free PMC article. Review.
-
Phosphorylation and subcellular localization of human phospholipase A1, DDHD1/PA-PLA1.Methods Enzymol. 2022;675:235-273. doi: 10.1016/bs.mie.2022.07.011. Epub 2022 Sep 2. Methods Enzymol. 2022. PMID: 36220272
-
The Spastic Paraplegia-Associated Phospholipase DDHD1 Is a Primary Brain Phosphatidylinositol Lipase.Biochemistry. 2018 Oct 2;57(39):5759-5767. doi: 10.1021/acs.biochem.8b00810. Epub 2018 Sep 17. Biochemistry. 2018. PMID: 30221923 Free PMC article.
-
Mutations in DDHD1, encoding a phospholipase A1, is a novel cause of retinopathy and neurodegeneration with brain iron accumulation.Eur J Med Genet. 2017 Dec;60(12):639-642. doi: 10.1016/j.ejmg.2017.08.015. Epub 2017 Aug 14. Eur J Med Genet. 2017. PMID: 28818478
-
Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family.Biochimie. 2007 Feb;89(2):197-204. doi: 10.1016/j.biochi.2006.09.021. Epub 2006 Oct 27. Biochimie. 2007. PMID: 17101204 Review.
Cited by
-
AS160 is a lipid-responsive regulator of cardiac Ca2+ homeostasis by controlling lysophosphatidylinositol metabolism and signaling.Nat Commun. 2024 Nov 6;15(1):9602. doi: 10.1038/s41467-024-54031-5. Nat Commun. 2024. PMID: 39505896 Free PMC article.
-
The function and mechanism of protein acylation in the regulation of viral infection.Virulence. 2025 Dec;16(1):2530171. doi: 10.1080/21505594.2025.2530171. Epub 2025 Jul 17. Virulence. 2025. PMID: 40673681 Free PMC article. Review.
-
From Classical to Alternative Pathways of 2-Arachidonoylglycerol Synthesis: AlterAGs at the Crossroad of Endocannabinoid and Lysophospholipid Signaling.Molecules. 2024 Aug 4;29(15):3694. doi: 10.3390/molecules29153694. Molecules. 2024. PMID: 39125098 Free PMC article. Review.
-
Current Knowledge on Mammalian Phospholipase A1, Brief History, Structures, Biochemical and Pathophysiological Roles.Molecules. 2022 Apr 12;27(8):2487. doi: 10.3390/molecules27082487. Molecules. 2022. PMID: 35458682 Free PMC article. Review.
-
A Light-Activatable Photocaged Variant of the Ultra-High Affinity ALFA-Tag Nanobody.Chembiochem. 2022 Jun 20;23(12):e202200079. doi: 10.1002/cbic.202200079. Epub 2022 Apr 27. Chembiochem. 2022. PMID: 35411584 Free PMC article.
References
-
- Waite M. vol 5. Plenum; New York, NY: 1987. pp. 1–332. (The Phospholipases: Handbook of Lipid Research).
-
- Pete M.J., Ross A.H., Exton J.H. Purification and properties of phospholipase A1 from bovine brain. J. Biol. Chem. 1994;269:19494–19500. - PubMed
-
- Yamashita A., Oka S., Tanikawa T., Hayashi Y., Nemoto-Sasaki Y., Sugiura T. The actions and metabolism of lysophosphatidylinositol, an endogenous agonist for GPR55. Prostaglandins Other Lipid Mediat. 2013;107:103–116. - PubMed
-
- Yamashita A., Hayashi Y., Nemoto-Sasaki Y., Ito M., Oka S., Tanikawa T., Waku K., Sugiura T. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 2014;53:18–81. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

