Proteomics-Based Insights Into the SARS-CoV-2-Mediated COVID-19 Pandemic: A Review of the First Year of Research
- PMID: 34089862
- PMCID: PMC8176883
- DOI: 10.1016/j.mcpro.2021.100103
Proteomics-Based Insights Into the SARS-CoV-2-Mediated COVID-19 Pandemic: A Review of the First Year of Research
Abstract
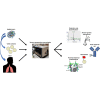
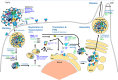
In late 2019, a virus subsequently named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in China and led to a worldwide pandemic of the disease termed coronavirus disease 2019. The global health threat posed by this pandemic led to an extremely rapid and robust mobilization of the scientific and medical communities as evidenced by the publication of more than 10,000 peer-reviewed articles and thousands of preprints in the first year of the pandemic alone. With the publication of the initial genome sequence of SARS-CoV-2, the proteomics community immediately joined this effort publishing, to date, more than 100 peer-reviewed proteomics studies and submitting many more preprints to preprint servers. In this review, we focus on peer-reviewed articles published on the proteome, glycoproteome, and glycome of SARS-CoV-2. At a basic level, proteomic studies provide valuable information on quantitative aspects of viral infection course; information on the identities, sites, and microheterogeneity of post-translational modifications; and, information on protein-protein interactions. At a biological systems level, these studies elucidate host cell and tissue responses, characterize antibodies and other immune system factors in infection, suggest biomarkers that may be useful for diagnosis and disease-course monitoring, and help in the development or repurposing of potential therapeutics. Here, we summarize results from selected early studies to provide a perspective on the current rapidly evolving literature.
Keywords: COVID-19; MS; SARS-CoV-2; glycosylation; review.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures


Similar articles
-
Proteomic Approaches to Study SARS-CoV-2 Biology and COVID-19 Pathology.J Proteome Res. 2021 Feb 5;20(2):1133-1152. doi: 10.1021/acs.jproteome.0c00764. Epub 2021 Jan 19. J Proteome Res. 2021. PMID: 33464917 Free PMC article.
-
SARS-CoV-2 spike protein receptor-binding domain N-glycans facilitate viral internalization in respiratory epithelial cells.Biochem Biophys Res Commun. 2021 Nov 19;579:69-75. doi: 10.1016/j.bbrc.2021.09.053. Epub 2021 Sep 23. Biochem Biophys Res Commun. 2021. PMID: 34592572 Free PMC article.
-
Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV.Nature. 2021 Jun;594(7862):246-252. doi: 10.1038/s41586-021-03493-4. Epub 2021 Apr 12. Nature. 2021. PMID: 33845483
-
Can Host Cell Proteins Like ACE2, ADAM17, TMPRSS2, Androgen Receptor be the Efficient Targets in SARS-CoV-2 Infection?Curr Drug Targets. 2021;22(10):1149-1157. doi: 10.2174/1389450121999201125201112. Curr Drug Targets. 2021. PMID: 33243116 Review.
-
SARS-CoV-2, ACE2 expression, and systemic organ invasion.Physiol Genomics. 2021 Feb 1;53(2):51-60. doi: 10.1152/physiolgenomics.00087.2020. Epub 2020 Dec 4. Physiol Genomics. 2021. PMID: 33275540 Free PMC article. Review.
Cited by
-
Protein Posttranslational Signatures Identified in COVID-19 Patient Plasma.Front Cell Dev Biol. 2022 Feb 11;10:807149. doi: 10.3389/fcell.2022.807149. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35223838 Free PMC article.
-
Proteomic insights into SARS-CoV-2 infection mechanisms, diagnosis, therapies and prognostic monitoring methods.Front Immunol. 2022 Sep 20;13:923387. doi: 10.3389/fimmu.2022.923387. eCollection 2022. Front Immunol. 2022. PMID: 36203586 Free PMC article. Review.
-
IFITM3, FURIN, ACE1, and TNF-α Genetic Association With COVID-19 Outcomes: Systematic Review and Meta-Analysis.Front Genet. 2022 Apr 1;13:775246. doi: 10.3389/fgene.2022.775246. eCollection 2022. Front Genet. 2022. PMID: 35432458 Free PMC article.
-
Instrumentation at the Leading Edge of Proteomics.Anal Chem. 2024 May 21;96(20):7976-8010. doi: 10.1021/acs.analchem.3c04497. Epub 2024 May 13. Anal Chem. 2024. PMID: 38738990 Free PMC article. Review.
-
Evaluation of serum and urine biomarkers for severe COVID-19.Front Med (Lausanne). 2024 Mar 6;11:1357659. doi: 10.3389/fmed.2024.1357659. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38510452 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

