Utilizing MIKC-type MADS-box protein SOC1 for yield potential enhancement in maize
- PMID: 34091722
- PMCID: PMC8376726
- DOI: 10.1007/s00299-021-02722-4
Utilizing MIKC-type MADS-box protein SOC1 for yield potential enhancement in maize
Abstract
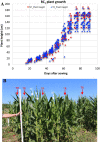
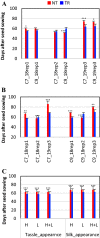
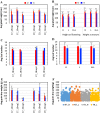
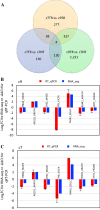
Overexpression of Zea mays SOC gene promotes flowering, reduces plant height, and leads to no reduction in grain production per plant, suggesting enhanced yield potential, at least, through increasing planting density. MIKC-type MADS-box gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) is an integrator conserved in the plant flowering pathway. In this study, the maize SOC1 (ZmSOC1) gene was cloned and overexpressed in transgenic maize Hi-II genotype. The T0 plants were backcrossed with nontransgenic inbred B73 to produce first generation backcross (BC1) seeds. Phenotyping of both transgenic and null segregant (NT) BC1 plants was conducted in three independent experiments. The BC1 transgenic plants showed new attributes such as increased vegetative growth, accelerated flowering time, reduced overall plant height, and increased grain weight. Second generation backcross (BC2) plants were evaluated in the field using two planting densities. Compared to BC2 NT plants, BC2 transgenic plants, were 12-18% shorter, flowered 5 days earlier, and showed no reduction in grain production per plant and an increase in fat, starch, and simple sugars in the grain. Transcriptome comparison in young leaves of 56-day-old BC1 plants revealed that the overexpressed ZmSOC1 resulted in 107 differentially expressed genes. The upregulated transcription factor DNA BINDING WITH ONE FINGER 5.4 (DOF5.4) was among the genes responsible for the reduced plant height. Modulating expression of SOC1 opens a new and effective approach to promote flowering and reduce plant height, which may have potential to enhance crop yield and improve grain quality.
Keywords: Dwarf plant; Flowering; SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1; Zea mays.
© 2021. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
Similar articles
-
Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering.Sci Rep. 2021 Jun 17;11(1):12758. doi: 10.1038/s41598-021-92215-x. Sci Rep. 2021. PMID: 34140602 Free PMC article.
-
Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco.Plant Cell Rep. 2013 Nov;32(11):1819-26. doi: 10.1007/s00299-013-1495-1. Epub 2013 Aug 21. Plant Cell Rep. 2013. PMID: 23963585
-
Flowering Time-Regulated Genes in Maize Include the Transcription Factor ZmMADS1.Plant Physiol. 2016 Sep;172(1):389-404. doi: 10.1104/pp.16.00285. Epub 2016 Jul 25. Plant Physiol. 2016. PMID: 27457125 Free PMC article.
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
-
Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: expectations and unanticipated outcomes.Plant Sci. 2013 Oct;211:52-60. doi: 10.1016/j.plantsci.2013.06.009. Epub 2013 Jul 12. Plant Sci. 2013. PMID: 23987811 Review.
Cited by
-
Constitutive expression of full-length or partial of SOC1 genes for yield enhancement in tomato.Front Plant Sci. 2025 Jul 28;16:1640731. doi: 10.3389/fpls.2025.1640731. eCollection 2025. Front Plant Sci. 2025. PMID: 40791786 Free PMC article.
-
Identification and expression analysis of the MADS-box genes of Kentucky bluegrass during inflorescence development.Physiol Mol Biol Plants. 2022 Jul;28(7):1359-1374. doi: 10.1007/s12298-022-01216-1. Epub 2022 Aug 22. Physiol Mol Biol Plants. 2022. PMID: 36051235 Free PMC article.
-
Genome wide association analysis for yield related traits in maize.BMC Plant Biol. 2022 Sep 21;22(1):449. doi: 10.1186/s12870-022-03812-5. BMC Plant Biol. 2022. PMID: 36127632 Free PMC article.
-
Regulatory frameworks involved in the floral induction, formation and developmental programming of woody horticultural plants: a case study on blueberries.Front Plant Sci. 2024 Feb 12;15:1336892. doi: 10.3389/fpls.2024.1336892. eCollection 2024. Front Plant Sci. 2024. PMID: 38410737 Free PMC article. Review.
-
zmm28 transgenic maize increases both N uptake- and N utilization-efficiencies.Commun Biol. 2022 Jun 7;5(1):555. doi: 10.1038/s42003-022-03501-x. Commun Biol. 2022. PMID: 35672405 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

