Perturbed hematopoiesis in individuals with germline DNMT3A overgrowth Tatton-Brown-Rahman syndrome
- PMID: 34092059
- PMCID: PMC8968878
- DOI: 10.3324/haematol.2021.278990
Perturbed hematopoiesis in individuals with germline DNMT3A overgrowth Tatton-Brown-Rahman syndrome
Abstract
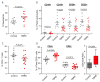
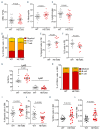
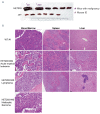
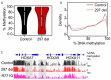
Tatton-Brown-Rahman syndrome (TBRS) is an overgrowth disorder caused by germline heterozygous mutations in the DNA methyltransferase DNMT3A. DNMT3A is a critical regulator of hematopoietic stem cell (HSC) differentiation and somatic DNMT3A mutations are frequent in hematologic malignancies and clonal hematopoiesis. Yet, the impact of constitutive DNMT3A mutation on hematopoiesis in TBRS is undefined. In order to establish how constitutive mutation of DNMT3A impacts blood development in TBRS we gathered clinical data and analyzed blood parameters in 18 individuals with TBRS. We also determined the distribution of major peripheral blood cell lineages by flow cytometric analyses. Our analyses revealed non-anemic macrocytosis, a relative decrease in lymphocytes and increase in neutrophils in TBRS individuals compared to unaffected controls. We were able to recapitulate these hematologic phenotypes in multiple murine models of TBRS and identified rare hematological and non-hematological malignancies associated with constitutive Dnmt3a mutation. We further show that loss of DNMT3A in TBRS is associated with an altered DNA methylation landscape in hematopoietic cells affecting regions critical to stem cell function and tumorigenesis. Overall, our data identify key hematopoietic effects driven by DNMT3A mutation with clinical implications for individuals with TBRS and DNMT3A-associated clonal hematopoiesis or malignancies.
Figures



Similar articles
-
The spectrum of DNMT3A variants in Tatton-Brown-Rahman syndrome overlaps with that in hematologic malignancies.Am J Med Genet A. 2017 Nov;173(11):3022-3028. doi: 10.1002/ajmg.a.38485. Epub 2017 Sep 21. Am J Med Genet A. 2017. PMID: 28941052
-
Skeletal abnormalities in mice with Dnmt3a missense mutations.Bone. 2024 Jun;183:117085. doi: 10.1016/j.bone.2024.117085. Epub 2024 Mar 23. Bone. 2024. PMID: 38522809 Free PMC article.
-
Acute myeloid leukaemia in a case with Tatton-Brown-Rahman syndrome: the peculiar DNMT3A R882 mutation.J Med Genet. 2017 Dec;54(12):805-808. doi: 10.1136/jmedgenet-2017-104574. Epub 2017 Apr 21. J Med Genet. 2017. PMID: 28432085 Review.
-
Expanding the genetic and clinical spectrum of Tatton-Brown-Rahman syndrome in a series of 24 French patients.J Med Genet. 2024 Aug 29;61(9):878-885. doi: 10.1136/jmg-2024-110031. J Med Genet. 2024. PMID: 38937076
-
Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown-Rahman overgrowth syndrome as a constitutional mutation.Am J Med Genet A. 2017 Jan;173(1):250-253. doi: 10.1002/ajmg.a.37995. Epub 2016 Nov 7. Am J Med Genet A. 2017. PMID: 27991732 Review.
Cited by
-
DNA Methylation in the Hypothalamic Feeding Center and Obesity.J Obes Metab Syndr. 2023 Dec 30;32(4):303-311. doi: 10.7570/jomes23073. Epub 2023 Dec 21. J Obes Metab Syndr. 2023. PMID: 38124554 Free PMC article. Review.
-
Distinct disease mutations in DNMT3A result in a spectrum of behavioral, epigenetic, and transcriptional deficits.bioRxiv [Preprint]. 2023 Feb 27:2023.02.27.530041. doi: 10.1101/2023.02.27.530041. bioRxiv. 2023. Update in: Cell Rep. 2023 Nov 28;42(11):113411. doi: 10.1016/j.celrep.2023.113411. PMID: 36909558 Free PMC article. Updated. Preprint.
-
Methylation signatures in clinically variable syndromic disorders: a familial DNMT3A variant in two adults with Tatton-Brown-Rahman syndrome.Eur J Hum Genet. 2023 Dec;31(12):1350-1354. doi: 10.1038/s41431-023-01459-w. Epub 2023 Sep 22. Eur J Hum Genet. 2023. PMID: 37736838 Free PMC article. No abstract available.
-
Constitutive loss of DNMT3A causes morbid obesity through misregulation of adipogenesis.Elife. 2022 May 30;11:e72359. doi: 10.7554/eLife.72359. Elife. 2022. PMID: 35635747 Free PMC article.
-
DNMT3A regulates murine megakaryocyte-biased hematopoietic stem cell fate decisions.Blood Adv. 2025 May 13;9(9):2285-2299. doi: 10.1182/bloodadvances.2024015061. Blood Adv. 2025. PMID: 40048738 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

