Preliminary Characterization of Two Small Insulinase-Like Proteases in Cryptosporidium parvum
- PMID: 34093467
- PMCID: PMC8175991
- DOI: 10.3389/fmicb.2021.651512
Preliminary Characterization of Two Small Insulinase-Like Proteases in Cryptosporidium parvum
Abstract
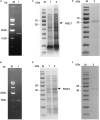
Cryptosporidium parvum is a major cause of moderate-to-severe diarrhea in humans and animals. Its compact genome contains 22 genes encoding divergent insulinase-like proteases (INS), which are poorly characterized. In this study, two small members of this family, INS-21 encoded by cgd7_2080 and INS-23 encoded by cgd5_3400, were cloned, expressed, and characterized to understand their functions. Recombinant INS-21 and INS-23 were expressed in Escherichia coli and polyclonal antibodies against these two proteins were prepared. The cgd7_2080 gene had a high transcription level during 0-2 h of in vitro C. parvum culture, while cgd5_3400 was highly transcribed at 0-6 h. INS-21 was mostly located in the apical region of sporozoites and merozoites whereas INS-23 was found as spots in sporozoites and merozoites. The immunoelectron microscopy confirmed the expression of INS-21 in the apical region of sporozoites while INS-23 appeared to be expressed in the dense granules of sporozoites. The neutralization efficiency was approximately 35%, when the cultures were treated with anti-INS23 antibodies. These results suggest that INS-21 and INS-23 are expressed in different organelles and might have different functions in the development of C. parvum.
Keywords: Cryptosporidium parvum; expression; insulinase-like protease; invasion; localization.
Copyright © 2021 Xu, Lai, Yang, Zhang, Li, Guo, Xiao and Feng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


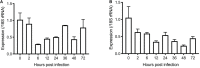


Similar articles
-
Characterization of Dense Granule Metalloproteinase INS-16 in Cryptosporidium parvum.Int J Mol Sci. 2022 Jul 10;23(14):7617. doi: 10.3390/ijms23147617. Int J Mol Sci. 2022. PMID: 35886965 Free PMC article.
-
Characterization of INS-15, A Metalloprotease Potentially Involved in the Invasion of Cryptosporidium parvum.Microorganisms. 2019 Oct 14;7(10):452. doi: 10.3390/microorganisms7100452. Microorganisms. 2019. PMID: 31615118 Free PMC article.
-
Expression and Functional Studies of INS-5, an Insulinase-Like Protein in Cryptosporidium parvum.Front Microbiol. 2020 May 8;11:719. doi: 10.3389/fmicb.2020.00719. eCollection 2020. Front Microbiol. 2020. PMID: 32457703 Free PMC article.
-
Comparative Study of Two Insulinlike Proteases in Cryptosporidium parvum.Microorganisms. 2021 Apr 16;9(4):861. doi: 10.3390/microorganisms9040861. Microorganisms. 2021. PMID: 33923793 Free PMC article.
-
Characterization of MEDLE-1, a protein in early development of Cryptosporidium parvum.Parasit Vectors. 2018 May 23;11(1):312. doi: 10.1186/s13071-018-2889-2. Parasit Vectors. 2018. PMID: 29792229 Free PMC article.
Cited by
-
Stage-specific expression and divergent functions of two insulinase-like proteases associated with host infectivity in Cryptosporidium.PLoS Negl Trop Dis. 2025 Jan 13;19(1):e0012777. doi: 10.1371/journal.pntd.0012777. eCollection 2025 Jan. PLoS Negl Trop Dis. 2025. PMID: 39804945 Free PMC article.
-
Characterization of Dense Granule Metalloproteinase INS-16 in Cryptosporidium parvum.Int J Mol Sci. 2022 Jul 10;23(14):7617. doi: 10.3390/ijms23147617. Int J Mol Sci. 2022. PMID: 35886965 Free PMC article.
-
Involvement of INS15 in the development and pathogenicity of the zoonotic pathogen Cryptosporidium parvum.PLoS Negl Trop Dis. 2024 Oct 3;18(10):e0012569. doi: 10.1371/journal.pntd.0012569. eCollection 2024 Oct. PLoS Negl Trop Dis. 2024. PMID: 39361715 Free PMC article.
-
Calcium-dependent protein kinases 2A involved in the growth of both asexual and sexual stages of Cryptosporidium parvum.PLoS Negl Trop Dis. 2025 May 28;19(5):e0013107. doi: 10.1371/journal.pntd.0013107. eCollection 2025 May. PLoS Negl Trop Dis. 2025. PMID: 40435438 Free PMC article.
-
Characterization of CpCaM, a protein potentially involved in the growth of Cryptosporidium parvum.Parasitol Res. 2023 Apr;122(4):989-996. doi: 10.1007/s00436-023-07803-9. Epub 2023 Mar 7. Parasitol Res. 2023. PMID: 36879147
References
LinkOut - more resources
Full Text Sources

