Understanding How Staphylococcal Autolysin Domains Interact With Polystyrene Surfaces
- PMID: 34093472
- PMCID: PMC8170090
- DOI: 10.3389/fmicb.2021.658373
Understanding How Staphylococcal Autolysin Domains Interact With Polystyrene Surfaces
Abstract
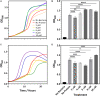
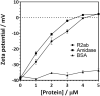
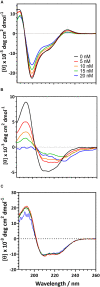
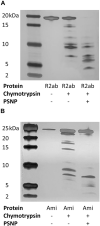
Biofilms, when formed on medical devices, can cause malfunctions and reduce the efficiency of these devices, thus complicating treatments and serving as a source of infection. The autolysin protein of Staphylococcus epidermidis contributes to its biofilm forming ability, especially on polystyrene surfaces. R2ab and amidase are autolysin protein domains thought to have high affinity to polystyrene surfaces, and they are involved in initial bacterial attachment in S. epidermidis biofilm formation. However, the structural details of R2ab and amidase binding to surfaces are poorly understood. In this study, we have investigated how R2ab and amidase influence biofilm formation on polystyrene surfaces. We have also studied how these proteins interact with polystyrene nanoparticles (PSNPs) using biophysical techniques. Pretreating polystyrene plates with R2ab and amidase domains inhibits biofilm growth relative to a control protein, indicating that these domains bind tightly to polystyrene surfaces and can block bacterial attachment. Correspondingly, we find that both domains interact strongly with anionic, carboxylate-functionalized as well as neutral, non-functionalized PSNPs, suggesting a similar binding interaction for nanoparticles and macroscopic surfaces. Both anionic and neutral PSNPs induce changes to the secondary structure of both R2ab and amidase as monitored by circular dichroism (CD) spectroscopy. These changes are very similar, though not identical, for both types of PSNPs, suggesting that carboxylate functionalization is only a small perturbation for R2ab and amidase binding. This structural change is also seen in limited proteolysis experiments, which exhibit substantial differences for both proteins when in the presence of carboxylate PSNPs. Overall, our results demonstrate that the R2ab and amidase domains strongly favor adsorption to polystyrene surfaces, and that surface adsorption destabilizes the secondary structure of these domains. Bacterial attachment to polystyrene surfaces during the initial phases of biofilm formation, therefore, may be mediated by aromatic residues, since these residues are known to drive adsorption to PSNPs. Together, these experiments can be used to develop new strategies for biofilm eradication, ensuring the proper long-lived functioning of medical devices.
Keywords: autolysin proteins; biofilms; medical implants; polystyrene nanoparticles; surface chemistry.
Copyright © 2021 Somarathne, Chappell, Perera, Yadav, Park and Fitzkee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Andrade J. D., Hlady V. (1986). “Protein adsorption and materials biocompatibility: a tutorial review and suggested hypotheses,” in Biopolymers/Non-Exclusion HPLC, ed. Dusek K. (Berlin: Spinger-Verlag; ), 10.1007/3-540-16422-7_6 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

