Positioning Diverse Type IV Structures and Functions Within Class 1 CRISPR-Cas Systems
- PMID: 34093491
- PMCID: PMC8175902
- DOI: 10.3389/fmicb.2021.671522
Positioning Diverse Type IV Structures and Functions Within Class 1 CRISPR-Cas Systems
Abstract
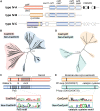
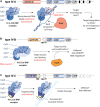
Type IV CRISPR systems encode CRISPR associated (Cas)-like proteins that combine with small RNAs to form multi-subunit ribonucleoprotein complexes. However, the lack of Cas nucleases, integrases, and other genetic features commonly observed in most CRISPR systems has made it difficult to predict type IV mechanisms of action and biological function. Here we summarize recent bioinformatic and experimental advancements that collectively provide the first glimpses into the function of specific type IV subtypes. We also provide a bioinformatic and structural analysis of type IV-specific proteins within the context of multi-subunit (class 1) CRISPR systems, informing future studies aimed at elucidating the function of these cryptic systems.
Keywords: CRISPR; Cas; Cas6; Cas7; CysH; DinG helicase; type IV.
Copyright © 2021 Taylor, Laderman, Armbrust, Hallmark, Keiser, Bondy-Denomy and Jackson.
Conflict of interest statement
JB-D is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics, and a scientific advisory board member and co-founder of Acrigen Biosciences. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.Nat Microbiol. 2019 Jan;4(1):89-96. doi: 10.1038/s41564-018-0274-8. Epub 2018 Nov 5. Nat Microbiol. 2019. PMID: 30397343
-
Structure of a type IV CRISPR-Cas ribonucleoprotein complex.iScience. 2021 Feb 17;24(3):102201. doi: 10.1016/j.isci.2021.102201. eCollection 2021 Mar 19. iScience. 2021. PMID: 33733066 Free PMC article.
-
Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease.RNA Biol. 2019 Oct;16(10):1438-1447. doi: 10.1080/15476286.2019.1634965. Epub 2019 Jun 28. RNA Biol. 2019. PMID: 31232162 Free PMC article.
-
DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes.FEMS Microbiol Rev. 2015 May;39(3):442-63. doi: 10.1093/femsre/fuv019. Epub 2015 Apr 30. FEMS Microbiol Rev. 2015. PMID: 25934119 Free PMC article. Review.
-
Approaches to study CRISPR RNA biogenesis and the key players involved.Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review.
Cited by
-
Evolution of Type IV CRISPR-Cas Systems: Insights from CRISPR Loci in Integrative Conjugative Elements of Acidithiobacillia.CRISPR J. 2021 Oct;4(5):656-672. doi: 10.1089/crispr.2021.0051. Epub 2021 Sep 28. CRISPR J. 2021. PMID: 34582696 Free PMC article.
-
CasDinG is a 5'-3' dsDNA and RNA/DNA helicase with three accessory domains essential for type IV CRISPR immunity.Nucleic Acids Res. 2023 Aug 25;51(15):8115-8132. doi: 10.1093/nar/gkad546. Nucleic Acids Res. 2023. PMID: 37395408 Free PMC article.
-
Structural variation of types IV-A1- and IV-A3-mediated CRISPR interference.Nat Commun. 2024 Oct 29;15(1):9306. doi: 10.1038/s41467-024-53778-1. Nat Commun. 2024. PMID: 39468082 Free PMC article.
-
Characterization of the self-targeting Type IV CRISPR interference system in Pseudomonas oleovorans.Nat Microbiol. 2022 Nov;7(11):1870-1878. doi: 10.1038/s41564-022-01229-2. Epub 2022 Sep 29. Nat Microbiol. 2022. PMID: 36175516
-
Insights into the Mechanism of CRISPR/Cas9-Based Genome Editing from Molecular Dynamics Simulations.ACS Omega. 2022 Dec 30;8(2):1817-1837. doi: 10.1021/acsomega.2c05583. eCollection 2023 Jan 17. ACS Omega. 2022. PMID: 36687047 Free PMC article. Review.

