Genomic Epidemiology of SARS-CoV-2 From Mainland China With Newly Obtained Genomes From Henan Province
- PMID: 34093495
- PMCID: PMC8172800
- DOI: 10.3389/fmicb.2021.673855
Genomic Epidemiology of SARS-CoV-2 From Mainland China With Newly Obtained Genomes From Henan Province
Abstract
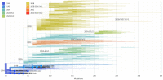
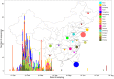
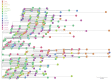
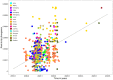
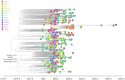
Even though the COVID-19 epidemic in China has been successfully put under control within a few months, it is still very important to infer the origin time and genetic diversity from the perspective of the whole genome sequence of its agent, SARS-CoV-2. Yet, the sequence of the entire virus genome from China in the current public database is very unevenly distributed with reference to time and place of collection. In particular, only one sequence was obtained in Henan province, adjacent to China's worst-case province, Hubei Province. Herein, we used high-throughput sequencing techniques to get 19 whole-genome sequences of SARS-CoV-2 from 18 severe patients admitted to the First Affiliated Hospital of Zhengzhou University, a provincial designated hospital for the treatment of severe COVID-19 cases in Henan province. The demographic, baseline, and clinical characteristics of these patients were described. To investigate the molecular epidemiology of SARS-CoV-2 of the current COVID-19 outbreak in China, 729 genome sequences (including 19 sequences from this study) sampled from Mainland China were analyzed with state-of-the-art comprehensive methods, including likelihood-mapping, split network, ML phylogenetic, and Bayesian time-scaled phylogenetic analyses. We estimated that the evolutionary rate and the time to the most recent common ancestor (TMRCA) of SARS-CoV-2 from Mainland China were 9.25 × 10-4 substitutions per site per year (95% BCI: 6.75 × 10-4 to 1.28 × 10-3) and October 1, 2019 (95% BCI: August 22, 2019 to November 6, 2019), respectively. Our results contribute to studying the molecular epidemiology and genetic diversity of SARS-CoV-2 over time in Mainland China.
Keywords: Henan Province; Mainland China; SARS-CoV-2; evolutionary rate; tMRCA; whole-genome sequence.
Copyright © 2021 Song, Cui and Zeng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Ferreira M. R., Suchard M.A. (2008). Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 36, 355–368. 10.1002/cjs.5550360302 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

