Identifying Genes Devoted to the Cell Death Process in the Gene Regulatory Network of Ustilago maydis
- PMID: 34093501
- PMCID: PMC8175908
- DOI: 10.3389/fmicb.2021.680290
Identifying Genes Devoted to the Cell Death Process in the Gene Regulatory Network of Ustilago maydis
Abstract
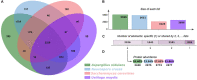
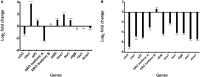
Cell death is a process that can be divided into three morphological patterns: apoptosis, autophagy and necrosis. In fungi, cell death is induced in response to intracellular and extracellular perturbations, such as plant defense molecules, toxins and fungicides, among others. Ustilago maydis is a dimorphic fungus used as a model for pathogenic fungi of animals, including humans, and plants. Here, we reconstructed the transcriptional regulatory network of U. maydis, through homology inferences by using as templates the well-known gene regulatory networks (GRNs) of Saccharomyces cerevisiae, Aspergillus nidulans and Neurospora crassa. Based on this GRN, we identified transcription factors (TFs) as hubs and functional modules and calculated diverse topological metrics. In addition, we analyzed exhaustively the module related to cell death, with 60 TFs and 108 genes, where diverse cell proliferation, mating-type switching and meiosis, among other functions, were identified. To determine the role of some of these genes, we selected a set of 11 genes for expression analysis by qRT-PCR (sin3, rlm1, aif1, tdh3 [isoform A], tdh3 [isoform B], ald4, mca1, nuc1, tor1, ras1, and atg8) whose homologues in other fungi have been described as central in cell death. These genes were identified as downregulated at 72 h, in agreement with the beginning of the cell death process. Our results can serve as the basis for the study of transcriptional regulation, not only of the cell death process but also of all the cellular processes of U. maydis.
Keywords: U. maydis; apoptosis; autophagy; cell death; necrosis; regulatory networks; transcription factors.
Copyright © 2021 Soberanes-Gutiérrez, Pérez-Rueda, Ruíz-Herrera and Galán-Vásquez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Gene Regulatory Network Inference and Gene Module Regulating Virulence in Fusarium oxysporum.Front Microbiol. 2022 Jun 2;13:861528. doi: 10.3389/fmicb.2022.861528. eCollection 2022. Front Microbiol. 2022. PMID: 35722316 Free PMC article.
-
Regulation of Ustilago maydis dimorphism, sporulation, and pathogenic development by a transcription factor with a highly conserved APSES domain.Mol Plant Microbe Interact. 2010 Feb;23(2):211-22. doi: 10.1094/MPMI-23-2-0211. Mol Plant Microbe Interact. 2010. PMID: 20064064
-
Induction of apoptosis-like cell death and clearance of stress-induced intracellular protein aggregates: dual roles for Ustilago maydis metacaspase Mca1.Mol Microbiol. 2017 Dec;106(5):815-831. doi: 10.1111/mmi.13848. Epub 2017 Oct 16. Mol Microbiol. 2017. PMID: 28941233
-
Endocytosis in the plant-pathogenic fungus Ustilago maydis.Protoplasma. 2005 Oct;226(1-2):75-80. doi: 10.1007/s00709-005-0109-3. Epub 2005 Oct 20. Protoplasma. 2005. PMID: 16231103 Review.
-
Ustilago maydis: how its biology relates to pathogenic development.New Phytol. 2004 Oct;164(1):31-42. doi: 10.1111/j.1469-8137.2004.01156.x. New Phytol. 2004. PMID: 33873482 Review.
Cited by
-
Construction and analysis of gene co-expression network in the pathogenic fungus Ustilago maydis.Front Microbiol. 2022 Dec 7;13:1048694. doi: 10.3389/fmicb.2022.1048694. eCollection 2022. Front Microbiol. 2022. PMID: 36569046 Free PMC article.
-
Gene Regulatory Network Inference and Gene Module Regulating Virulence in Fusarium oxysporum.Front Microbiol. 2022 Jun 2;13:861528. doi: 10.3389/fmicb.2022.861528. eCollection 2022. Front Microbiol. 2022. PMID: 35722316 Free PMC article.
-
A comprehensive transcription factor and DNA-binding motif resource for the construction of gene regulatory networks in Botrytis cinerea and Trichoderma atroviride.Comput Struct Biotechnol J. 2021 Nov 18;19:6212-6228. doi: 10.1016/j.csbj.2021.11.012. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34900134 Free PMC article.
-
A landscape of gene regulation in the parasitic amoebozoa Entamoeba spp.PLoS One. 2022 Aug 1;17(8):e0271640. doi: 10.1371/journal.pone.0271640. eCollection 2022. PLoS One. 2022. PMID: 35913975 Free PMC article.
-
Homology-based reconstruction of regulatory networks for bacterial and archaeal genomes.Front Microbiol. 2022 Jul 19;13:923105. doi: 10.3389/fmicb.2022.923105. eCollection 2022. Front Microbiol. 2022. PMID: 35928164 Free PMC article.
References
-
- Agrios G. N. (2005). Environmental Effects on The Development of Infectious Plant Disease. Plant Pathology, 5th Edn. Cambridge, MA: Elsevier Academic Press, USA, 251–265.
-
- Aguilar L. R., Pardo J. P., Lomelí M. M., Bocardo O. I. L., Oropeza M. A. J., Sánchez G. G. (2017). Lipid droplets accumulation and other biochemical changes induced in the fungal pathogen Ustilago maydis under nitrogen-starvation. Arch. Microbiol. 199 1195–1209. 10.1007/s00203-017-1388-8 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

