A Nitric Oxide-Responsive Transcriptional Regulator NsrR Cooperates With Lrp and CRP to Tightly Control the hmpA Gene in Vibrio vulnificus
- PMID: 34093504
- PMCID: PMC8175989
- DOI: 10.3389/fmicb.2021.681196
A Nitric Oxide-Responsive Transcriptional Regulator NsrR Cooperates With Lrp and CRP to Tightly Control the hmpA Gene in Vibrio vulnificus
Abstract
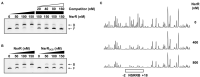
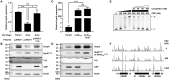
Nitric oxide (NO) is an important antimicrobial effector produced by the host innate immune system to counteract invading pathogens. To survive and establish a successful infection, a fulminating human pathogen Vibrio vulnificus expresses the hmpA gene encoding an NO dioxygenase in an NO-responsive manner. In this study, we identified an Rrf2-family transcriptional regulator NsrR that is predicted to contain the Fe-S cluster coordinated by three cysteine residues. Transcriptome analysis showed that NsrR controls the expression of multiple genes potentially involved in nitrosative stress responses. Particularly, NsrR acts as a strong repressor of hmpA transcription and relieves the repression of hmpA upon exposure to NO. Notably, nsrR and hmpA are transcribed divergently, and their promoter regions overlap with each other. Molecular biological analyses revealed that NsrR directly binds to this overlapping promoter region, which is alleviated by loss of the Fe-S cluster, leading to the subsequent derepression of hmpA under nitrosative stress. We further found that a leucine-responsive regulatory protein (Lrp) negatively regulates hmpA in an NsrR-dependent manner by directly binding to the promoter region, presumably resulting in a DNA conformation change to support the repression by NsrR. Meanwhile, a cyclic AMP receptor protein (CRP) positively regulates hmpA probably through repression of nsrR and lrp by directly binding to each promoter region in a sequential cascade. Altogether, this collaborative regulation of NsrR along with Lrp and CRP enables an elaborate control of hmpA transcription, contributing to survival under host-derived nitrosative stress and thereby the pathogenesis of V. vulnificus.
Keywords: Vibrio vulnificus; gene regulation; nitric oxide; nitric oxide dioxygenase; nitrosative stress; stress response; transcriptional regulator.
Copyright © 2021 Choi, Kim, Im and Choi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

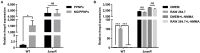




Similar articles
-
A MARTX Toxin rtxA Gene Is Controlled by Host Environmental Signals through a CRP-Coordinated Regulatory Network in Vibrio vulnificus.mBio. 2020 Jul 28;11(4):e00723-20. doi: 10.1128/mBio.00723-20. mBio. 2020. PMID: 32723914 Free PMC article.
-
The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator.J Bacteriol. 2006 Feb;188(3):874-81. doi: 10.1128/JB.188.3.874-881.2006. J Bacteriol. 2006. PMID: 16428390 Free PMC article.
-
NsrR from Streptomyces coelicolor is a nitric oxide-sensing [4Fe-4S] cluster protein with a specialized regulatory function.J Biol Chem. 2015 May 15;290(20):12689-704. doi: 10.1074/jbc.M115.643072. Epub 2015 Mar 14. J Biol Chem. 2015. PMID: 25771538 Free PMC article.
-
Genome-wide identification of binding sites for the nitric oxide-sensitive transcriptional regulator NsrR.Methods Enzymol. 2008;437:211-33. doi: 10.1016/S0076-6879(07)37012-2. Methods Enzymol. 2008. PMID: 18433631 Review.
-
The intricate workings of a bacterial epigenetic switch.Adv Exp Med Biol. 2004;547:83-9. doi: 10.1007/978-1-4419-8861-4_7. Adv Exp Med Biol. 2004. PMID: 15230094 Review.
Cited by
-
Transcriptomic Analysis Reveals That Municipal Wastewater Effluent Enhances Vibrio vulnificus Growth and Virulence Potential.Front Microbiol. 2021 Oct 25;12:754683. doi: 10.3389/fmicb.2021.754683. eCollection 2021. Front Microbiol. 2021. PMID: 34759904 Free PMC article.
-
S-Nitrosylation of the virulence regulator AphB promotes Vibrio cholerae pathogenesis.PLoS Pathog. 2022 Jun 17;18(6):e1010581. doi: 10.1371/journal.ppat.1010581. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35714156 Free PMC article.
-
Two novel genes identified by large-scale transcriptomic analysis are essential for biofilm and rugose colony development of Vibrio vulnificus.PLoS Pathog. 2023 Jan 19;19(1):e1011064. doi: 10.1371/journal.ppat.1011064. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36656902 Free PMC article.
-
Nitric Oxide Signaling for Aerial Mycelium Formation in Streptomyces coelicolor A3(2) M145.Appl Environ Microbiol. 2022 Dec 13;88(23):e0122222. doi: 10.1128/aem.01222-22. Epub 2022 Nov 10. Appl Environ Microbiol. 2022. PMID: 36354316 Free PMC article.
-
Quorum Sensing Coordinates Carbon and Nitrogen Metabolism to Optimize Public Goods Production in Pseudomonas fluorescens 2P24.Adv Sci (Weinh). 2025 Mar;12(12):e2412224. doi: 10.1002/advs.202412224. Epub 2025 Jan 31. Adv Sci (Weinh). 2025. PMID: 39888293 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

