Analysis of Porcine RIG-I Like Receptors Revealed the Positive Regulation of RIG-I and MDA5 by LGP2
- PMID: 34093517
- PMCID: PMC8169967
- DOI: 10.3389/fimmu.2021.609543
Analysis of Porcine RIG-I Like Receptors Revealed the Positive Regulation of RIG-I and MDA5 by LGP2
Abstract
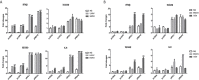
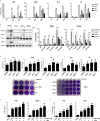
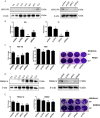
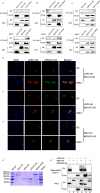
The RLRs play critical roles in sensing and fighting viral infections especially RNA virus infections. Despite the extensive studies on RLRs in humans and mice, there is a lack of systemic investigation of livestock animal RLRs. In this study, we characterized the porcine RLR members RIG-I, MDA5 and LGP2. Compared with their human counterparts, porcine RIG-I and MDA5 exhibited similar signaling activity to distinct dsRNA and viruses, via similar and cooperative recognitions. Porcine LGP2, without signaling activity, was found to positively regulate porcine RIG-I and MDA5 in transfected porcine alveolar macrophages (PAMs), gene knockout PAMs and PK-15 cells. Mechanistically, LGP2 interacts with RIG-I and MDA5 upon cell activation, and promotes the binding of dsRNA ligand by MDA5 as well as RIG-I. Accordingly, porcine LGP2 exerted broad antiviral functions. Intriguingly, we found that porcine LGP2 mutants with defects in ATPase and/or dsRNA binding present constitutive activity which are likely through RIG-I and MDA5. Our work provided significant insights into porcine innate immunity, species specificity and immune biology.
Keywords: LGP2; RIG-like receptors (RLRs); porcine; positive regulation; species specificity.
Copyright © 2021 Li, Yang, Zhu, Wang, Ji, Luo, Shao, Xu, Liu, Zheng, Meurens, Chen and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
LGP2 binds to PACT to regulate RIG-I- and MDA5-mediated antiviral responses.Sci Signal. 2019 Oct 1;12(601):eaar3993. doi: 10.1126/scisignal.aar3993. Sci Signal. 2019. PMID: 31575732
-
Presence of two RIG-I-like receptors, MDA5 and LGP2, and their dsRNA binding capacity in a perciform fish, the snakehead Channa argus.Dev Comp Immunol. 2022 Jan;126:104235. doi: 10.1016/j.dci.2021.104235. Epub 2021 Aug 18. Dev Comp Immunol. 2022. PMID: 34418428
-
MDA5 and LGP2 acts as a key regulator though activating NF-κB and IRF3 in RLRs signaling of mandarinfish.Fish Shellfish Immunol. 2019 Mar;86:1114-1122. doi: 10.1016/j.fsi.2018.12.054. Epub 2018 Dec 28. Fish Shellfish Immunol. 2019. PMID: 30594581
-
RIG-I-like receptors: Molecular mechanism of activation and signaling.Adv Immunol. 2023;158:1-74. doi: 10.1016/bs.ai.2023.03.001. Epub 2023 May 9. Adv Immunol. 2023. PMID: 37453753 Review.
-
LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review.
Cited by
-
Mammalian orthoreovirus capsid protein σ3 antagonizes RLR-mediated antiviral responses by degrading MAVS.mSphere. 2024 Jun 25;9(6):e0023624. doi: 10.1128/msphere.00236-24. Epub 2024 May 17. mSphere. 2024. PMID: 38757961 Free PMC article.
-
Development of Specific Monoclonal Antibodies against Porcine RIG-I-like Receptors Revealed the Species Specificity.Int J Mol Sci. 2023 Feb 18;24(4):4118. doi: 10.3390/ijms24044118. Int J Mol Sci. 2023. PMID: 36835527 Free PMC article.
-
Transcriptome analysis identifies LGP2 as an MDA5-mediated signaling activator following spring viremia of carp virus infection in common carp (Cyprinus carpio L.).Front Immunol. 2022 Oct 18;13:1019872. doi: 10.3389/fimmu.2022.1019872. eCollection 2022. Front Immunol. 2022. PMID: 36330521 Free PMC article.
-
Proteomic Characterization of PAMs with PRRSV-ADE Infection.Viruses. 2022 Dec 22;15(1):36. doi: 10.3390/v15010036. Viruses. 2022. PMID: 36680075 Free PMC article.
-
Chicken-Derived Pattern Recognition Receptor chLGP2 Inhibits the Replication and Proliferation of Infectious Bronchitis Virus.Front Microbiol. 2022 Jan 25;12:810215. doi: 10.3389/fmicb.2021.810215. eCollection 2021. Front Microbiol. 2022. PMID: 35145497 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

