Mitf Involved in Innate Immunity by Activating Tyrosinase-Mediated Melanin Synthesis in Pteria penguin
- PMID: 34093521
- PMCID: PMC8173187
- DOI: 10.3389/fimmu.2021.626493
Mitf Involved in Innate Immunity by Activating Tyrosinase-Mediated Melanin Synthesis in Pteria penguin
Abstract
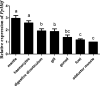
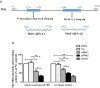
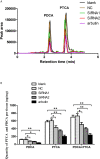
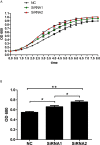
The microphthalmia-associated transcription factor (MITF) is an important transcription factor that plays a key role in melanogenesis, cell proliferation, survival and immune defense in vertebrate. However, its function and function mechanism in bivalve are still rarely known. In this research, first, a Mitf gene was characterized from Pteria penguin (P. penguin). The PpMitf contained an open reading frame of 1,350 bp, encoding a peptide of 449 deduced amino acids with a highly conserved basic helix-loop-helix-leucine zipper (bHLH-LZ) domain. The PpMITF shared 55.7% identity with amino acid sequence of Crassostrea gigas (C. gigas). Tissue distribution analysis revealed that PpMitf was highly expressed in mantle and hemocytes, which were important tissues for color formation and innate immunity. Second, the functions of PpMitf in melanin synthesis and innate immunity were identified. The PpMitf silencing significantly decreased the tyrosinase activity and melanin content, indicating PpMitf involved in melanin synthesis of P. penguin. Meanwhile, the PpMitf silencing clearly down-regulated the expression of PpBcl2 (B cell lymphoma/leukemia-2 gene) and antibacterial activity of hemolymph supernatant, indicating that PpMitf involved in innate immunity of P. penguin. Third, the function mechanism of PpMitf in immunity was analyzed. The promoter sequence analysis of tyrosinase (Tyr) revealed two highly conserved E-box elements, which were specifically recognized by HLH-LZ of MITF. The luciferase activities analysis showed that Mitf could activate the E-box in Tyr promoter through highly conserved bHLH-LZ domain, and demonstrated that PpMitf involved in melanin synthesis and innate immunity by regulating tyrosinase expression. Finally, melanin from P. penguin, the final production of Mitf-Tyr-melanin pathway, was confirmed to have direct antibacterial activity. The results collectively demonstrated that PpMitf played a key role in innate immunity through activating tyrosinase-mediated melanin synthesis in P. penguin.
Keywords: Mitf; Pteria penguin; innate immunity; melanin; tyrosinase.
Copyright © 2021 Yu, Lu, Zhong, Qu, Wang, Yu and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Creb2 involved in innate immunity by activating PpMitf-mediated melanogenesis in Pteria penguin.Fish Shellfish Immunol. 2023 Jul;138:108809. doi: 10.1016/j.fsi.2023.108809. Epub 2023 May 13. Fish Shellfish Immunol. 2023. PMID: 37182797
-
Pax3 Gene Regulated Melanin Synthesis by Tyrosinase Pathway in Pteria penguin.Int J Mol Sci. 2018 Nov 22;19(12):3700. doi: 10.3390/ijms19123700. Int J Mol Sci. 2018. PMID: 30469474 Free PMC article.
-
Mitf involved in shell pigmentation by activating tyrosinase-mediated melanin synthesis in Pacific oyster (Crassostrea gigas).Gene. 2024 Mar 1;897:148086. doi: 10.1016/j.gene.2023.148086. Epub 2023 Dec 15. Gene. 2024. PMID: 38104952
-
[Regulation of melanogenesis: the role of cAMP and MITF].Postepy Hig Med Dosw (Online). 2012 Jan 30;66:33-40. Postepy Hig Med Dosw (Online). 2012. PMID: 22371403 Review. Polish.
-
Fifteen-year quest for microphthalmia-associated transcription factor target genes.Pigment Cell Melanoma Res. 2010 Feb;23(1):27-40. doi: 10.1111/j.1755-148X.2009.00653.x. Epub 2009 Nov 25. Pigment Cell Melanoma Res. 2010. PMID: 19995375 Review.
Cited by
-
The lgi-miR-2d is Potentially Involved in Shell Melanin Synthesis by Targeting mitf in Manila Clam Ruditapes philippinarum.Mar Biotechnol (NY). 2024 Jun;26(3):432-446. doi: 10.1007/s10126-024-10307-x. Epub 2024 Apr 12. Mar Biotechnol (NY). 2024. PMID: 38607523
-
miRNA profiling of B16F10 melanoma cell exosomes reveals melanin synthesis-related genes.Heliyon. 2024 Apr 29;10(9):e30474. doi: 10.1016/j.heliyon.2024.e30474. eCollection 2024 May 15. Heliyon. 2024. PMID: 38711645 Free PMC article.
-
Hydrolyzed Feather Meal in Diet of Yellow Catfish (Pelteobagrus fulvidraco): Effects on Growth Performance, Flesh Quality, Skin Color, and Intestinal Flora.Aquac Nutr. 2025 Aug 31;2025:7200771. doi: 10.1155/anu/7200771. eCollection 2025. Aquac Nutr. 2025. PMID: 40923043 Free PMC article.
-
Anti-Melanogenic Effects of L-Theanine on B16F10 Cells and Zebrafish.Molecules. 2025 Feb 19;30(4):956. doi: 10.3390/molecules30040956. Molecules. 2025. PMID: 40005265 Free PMC article.
-
Enhanced insulin-regulated phagocytic activities support extreme health span and longevity in multiple populations.Aging Cell. 2023 May;22(5):e13810. doi: 10.1111/acel.13810. Epub 2023 Mar 8. Aging Cell. 2023. PMID: 36883688 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

