Study on the Immune Escape Mechanism of Acute Myeloid Leukemia With DNMT3A Mutation
- PMID: 34093541
- PMCID: PMC8173207
- DOI: 10.3389/fimmu.2021.653030
Study on the Immune Escape Mechanism of Acute Myeloid Leukemia With DNMT3A Mutation
Abstract
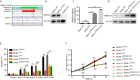
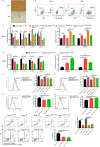
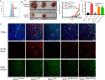
DNA (cytosine-5)-methyltransferase 3A (DNMT3A)-mutated acute myeloid leukemia (AML) has a poor prognosis, but the exact mechanism is still unclear. Here, we aimed to explore the mechanism of immune escape in AML with DNMT3A mutation. We constructed a DNMT3A knockout clone and DNMT3A-R882H-mutated clones. RNA-seq results showed that transcription factors and macrophage inflammatory proteins were significantly downregulated in the DNMT3A mutant clones. KEGG enrichment and gene set enrichment analysis (GSEA) showed that a large number of genes were enriched in inflammatory immune-related pathways, such as the toll-like receptor signaling pathway. Therefore, we co-cultured AML cells with macrophages. The DNMT3A-mutated AML cells attenuated M1 macrophage polarization and resisted its killing effect in vitro and in vivo. In xenografts, the tumor volumes in the experimental group were significantly larger than those in the control group, and the proportion of M2 macrophages was significantly higher. After the co-culture, the increase in pro-inflammatory cytokine expression in the mutant cells was significantly lower than that in the control group, while that in immunosuppressive factors was not significantly different. In co-cultivated supernatants, the concentration of inflammatory factors in the experimental group was significantly lower than that in the control group, while that of immunosuppressive factors was significantly higher. Resistin significantly promoted the expression of inflammatory proteins in AML cells. It relieved the inhibitory effect of DNMT3A mutation, promoted the phenotypic recovery of the co-cultured macrophages, eliminated resistance, and regulated the immune microenvironment. Thus, resistin may serve as an ancillary drug for patients with DNMT3A-mutated AML.
Keywords: AML; DNMT3A; epigenetics; immune escape; immune microenvironment; macrophage polarization.
Copyright © 2021 Que, Li, Lin, Zhu, Xiao, Wang, Zhu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

