Dynamics of DNA Methylation and Its Functions in Plant Growth and Development
- PMID: 34093600
- PMCID: PMC8175986
- DOI: 10.3389/fpls.2021.596236
Dynamics of DNA Methylation and Its Functions in Plant Growth and Development
Abstract
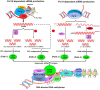
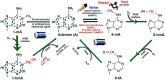
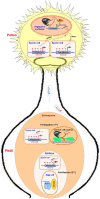
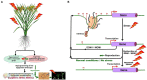
Epigenetic modifications in DNA bases and histone proteins play important roles in the regulation of gene expression and genome stability. Chemical modification of DNA base (e.g., addition of a methyl group at the fifth carbon of cytosine residue) switches on/off the gene expression during developmental process and environmental stresses. The dynamics of DNA base methylation depends mainly on the activities of the writer/eraser guided by non-coding RNA (ncRNA) and regulated by the developmental/environmental cues. De novo DNA methylation and active demethylation activities control the methylation level and regulate the gene expression. Identification of ncRNA involved in de novo DNA methylation, increased DNA methylation proteins guiding DNA demethylase, and methylation monitoring sequence that helps maintaining a balance between DNA methylation and demethylation is the recent developments that may resolve some of the enigmas. Such discoveries provide a better understanding of the dynamics/functions of DNA base methylation and epigenetic regulation of growth, development, and stress tolerance in crop plants. Identification of epigenetic pathways in animals, their existence/orthologs in plants, and functional validation might improve future strategies for epigenome editing toward climate-resilient, sustainable agriculture in this era of global climate change. The present review discusses the dynamics of DNA methylation (cytosine/adenine) in plants, its functions in regulating gene expression under abiotic/biotic stresses, developmental processes, and genome stability.
Keywords: 5-methylcytosine; DNA methylation; DNA modification; N6-methyladenine; environmental stress; epigenetics; gene regulation; plant growth.
Copyright © 2021 Kumar and Mohapatra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Epigenetic Modifications in Genome Help Remembering the Stress Tolerance Strategy Adopted by the Plant.Front Biosci (Landmark Ed). 2024 Mar 22;29(3):126. doi: 10.31083/j.fbl2903126. Front Biosci (Landmark Ed). 2024. PMID: 38538276 Review.
-
In Response to Abiotic Stress, DNA Methylation Confers EpiGenetic Changes in Plants.Plants (Basel). 2021 May 30;10(6):1096. doi: 10.3390/plants10061096. Plants (Basel). 2021. PMID: 34070712 Free PMC article. Review.
-
Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond.Front Genet. 2018 Dec 18;9:640. doi: 10.3389/fgene.2018.00640. eCollection 2018. Front Genet. 2018. PMID: 30619465 Free PMC article. Review.
-
Dynamics and function of DNA methylation in plants.Nat Rev Mol Cell Biol. 2018 Aug;19(8):489-506. doi: 10.1038/s41580-018-0016-z. Nat Rev Mol Cell Biol. 2018. PMID: 29784956 Review.
Cited by
-
Exploring distinct properties of endometrial stem cells through advanced single-cell analysis platforms.Stem Cell Res Ther. 2023 Dec 20;14(1):379. doi: 10.1186/s13287-023-03616-w. Stem Cell Res Ther. 2023. PMID: 38124100 Free PMC article. Review.
-
Arabidopsis thaliana Roots Exposed to Extracellular Self-DNA: Evidence of Epigenetic Effects.Epigenomes. 2025 Apr 30;9(2):13. doi: 10.3390/epigenomes9020013. Epigenomes. 2025. PMID: 40407422 Free PMC article.
-
Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review.Plants (Basel). 2024 Jul 27;13(15):2082. doi: 10.3390/plants13152082. Plants (Basel). 2024. PMID: 39124200 Free PMC article. Review.
-
ARABIDOPSIS HOMOLOG OF TRITHORAX1 impacts lateral root development by epigenetic regulation of targets involved in root system architecture.New Phytol. 2025 Sep;247(5):2180-2195. doi: 10.1111/nph.70349. Epub 2025 Jul 7. New Phytol. 2025. PMID: 40624794 Free PMC article.
-
Role of Epigenetics in Modulating Phenotypic Plasticity against Abiotic Stresses in Plants.Int J Genomics. 2022 Jun 14;2022:1092894. doi: 10.1155/2022/1092894. eCollection 2022. Int J Genomics. 2022. PMID: 35747076 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

