Large-Scale Multiplexing Permits Full-Length Transcriptome Annotation of 32 Bovine Tissues From a Single Nanopore Flow Cell
- PMID: 34093657
- PMCID: PMC8173071
- DOI: 10.3389/fgene.2021.664260
Large-Scale Multiplexing Permits Full-Length Transcriptome Annotation of 32 Bovine Tissues From a Single Nanopore Flow Cell
Abstract
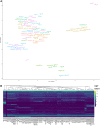
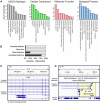
A comprehensive annotation of transcript isoforms in domesticated species is lacking. Especially considering that transcriptome complexity and splicing patterns are not well-conserved between species, this presents a substantial obstacle to genomic selection programs that seek to improve production, disease resistance, and reproduction. Recent advances in long-read sequencing technology have made it possible to directly extrapolate the structure of full-length transcripts without the need for transcript reconstruction. In this study, we demonstrate the power of long-read sequencing for transcriptome annotation by coupling Oxford Nanopore Technology (ONT) with large-scale multiplexing of 93 samples, comprising 32 tissues collected from adult male and female Hereford cattle. More than 30 million uniquely mapping full-length reads were obtained from a single ONT flow cell, and used to identify and characterize the expression dynamics of 99,044 transcript isoforms at 31,824 loci. Of these predicted transcripts, 21% exactly matched a reference transcript, and 61% were novel isoforms of reference genes, substantially increasing the ratio of transcript variants per gene, and suggesting that the complexity of the bovine transcriptome is comparable to that in humans. Over 7,000 transcript isoforms were extremely tissue-specific, and 61% of these were attributed to testis, which exhibited the most complex transcriptome of all interrogated tissues. Despite profiling over 30 tissues, transcription was only detected at about 60% of reference loci. Consequently, additional studies will be necessary to continue characterizing the bovine transcriptome in additional cell types, developmental stages, and physiological conditions. However, by here demonstrating the power of ONT sequencing coupled with large-scale multiplexing, the task of exhaustively annotating the bovine transcriptome - or any mammalian transcriptome - appears significantly more feasible.
Keywords: alternative splicyng; annotation; cattle; full-length transcript; long-read sequencing; tissue-specific; transcriptome.
Copyright © 2021 Halstead, Islas-Trejo, Goszczynski, Medrano, Zhou and Ross.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Prediction of transcript isoforms in 19 chicken tissues by Oxford Nanopore long-read sequencing.Front Genet. 2022 Oct 3;13:997460. doi: 10.3389/fgene.2022.997460. eCollection 2022. Front Genet. 2022. PMID: 36246588 Free PMC article.
-
A Comparative Full-Length Transcriptome Analysis Using Oxford Nanopore Technologies (ONT) in Four Tissues of Bovine Origin.Animals (Basel). 2024 May 31;14(11):1646. doi: 10.3390/ani14111646. Animals (Basel). 2024. PMID: 38891695 Free PMC article.
-
A de novo Full-Length mRNA Transcriptome Generated From Hybrid-Corrected PacBio Long-Reads Improves the Transcript Annotation and Identifies Thousands of Novel Splice Variants in Atlantic Salmon.Front Genet. 2021 Apr 27;12:656334. doi: 10.3389/fgene.2021.656334. eCollection 2021. Front Genet. 2021. PMID: 33986770 Free PMC article.
-
Methodologies for Transcript Profiling Using Long-Read Technologies.Front Genet. 2020 Jul 7;11:606. doi: 10.3389/fgene.2020.00606. eCollection 2020. Front Genet. 2020. PMID: 32733532 Free PMC article. Review.
-
Long-Read Sequencing - A Powerful Tool in Viral Transcriptome Research.Trends Microbiol. 2019 Jul;27(7):578-592. doi: 10.1016/j.tim.2019.01.010. Epub 2019 Feb 26. Trends Microbiol. 2019. PMID: 30824172 Review.
Cited by
-
Prediction of transcript isoforms in 19 chicken tissues by Oxford Nanopore long-read sequencing.Front Genet. 2022 Oct 3;13:997460. doi: 10.3389/fgene.2022.997460. eCollection 2022. Front Genet. 2022. PMID: 36246588 Free PMC article.
-
ICAnnoLncRNA: A Snakemake Pipeline for a Long Non-Coding-RNA Search and Annotation in Transcriptomic Sequences.Genes (Basel). 2023 Jun 24;14(7):1331. doi: 10.3390/genes14071331. Genes (Basel). 2023. PMID: 37510236 Free PMC article.
-
D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions.Cell Syst. 2021 Oct 20;12(10):969-982.e6. doi: 10.1016/j.cels.2021.08.010. Epub 2021 Oct 9. Cell Syst. 2021. PMID: 34536380 Free PMC article.
-
Comparison of Alternative Splicing Landscapes Revealed by Long-Read Sequencing in Hepatocyte-Derived HepG2 and Huh7 Cultured Cells and Human Liver Tissue.Biology (Basel). 2023 Dec 6;12(12):1494. doi: 10.3390/biology12121494. Biology (Basel). 2023. PMID: 38132320 Free PMC article.
-
The Research Progress of Single-Molecule Sequencing and Its Significance in Nucleic Acid Metrology.Biosensors (Basel). 2024 Dec 25;15(1):4. doi: 10.3390/bios15010004. Biosensors (Basel). 2024. PMID: 39852055 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

