Elevated Expression of PDZD11 Is Associated With Poor Prognosis and Immune Infiltrates in Hepatocellular Carcinoma
- PMID: 34093661
- PMCID: PMC8176286
- DOI: 10.3389/fgene.2021.669928
Elevated Expression of PDZD11 Is Associated With Poor Prognosis and Immune Infiltrates in Hepatocellular Carcinoma
Abstract
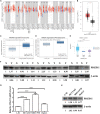
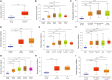
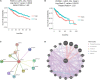
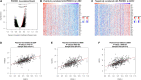
Epithelial cells are held together by tight and adherent junctions, which are destroyed by the activation of epithelial-to-mesenchymal transition (EMT). The PLEKHA7-PDZD11 complex has been reported to be important for epithelial cell adhesion and connecting tissues. However, there is no research regarding the expression and role of PDZD11 in liver hepatocellular carcinoma (LIHC) progression. Here, we analyzed PDZD11 mRNA expression and its clinical results in LIHC patient RNA sequencing data based on different open databases. Furthermore, we examined differences in PDZD11 expression in LIHC tissues and cell lines using western blotting and real-time qPCR. These results are the first to report that the mRNA and protein levels of PDZD11 are significantly overexpressed in LIHC. Moreover, high expression of PDZD11 was correlated with poor overall survival in patients with LIHC. Gene regulatory network analysis suggested that PDZD11 is mainly involved in copper ion homeostasis, proteasome, and oxidative phosphorylation pathways. Interestingly, we found that PDZD11 levels were positively correlated with the abundance of immune infiltrates. In particular, higher infiltration levels of CD4+ T cells and macrophage subsets significantly affected LIHC patient prognosis. Taken together, these results demonstrate that PDZD11 could be a potential diagnostic and prognostic biomarker in LIHC.
Keywords: PDZD11; functional network analysis; hepatocellular carcinoma; immune infiltrates; prognostic biomarker.
Copyright © 2021 Chen, Xie, Xie, Yang, Pang and Ye.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Evaluation of PDZD11 in hepatocellular carcinoma: prognostic value and diagnostic potential in combination with AFP.Front Oncol. 2025 Mar 25;15:1533865. doi: 10.3389/fonc.2025.1533865. eCollection 2025. Front Oncol. 2025. PMID: 40201341 Free PMC article.
-
Iron-sulphur cluster biogenesis factor LYRM4 is a novel prognostic biomarker associated with immune infiltrates in hepatocellular carcinoma.Cancer Cell Int. 2021 Sep 6;21(1):463. doi: 10.1186/s12935-021-02131-3. Cancer Cell Int. 2021. PMID: 34488769 Free PMC article.
-
CPNE1 is a potential prognostic biomarker, associated with immune infiltrates and promotes progression of hepatocellular carcinoma.Cancer Cell Int. 2022 Feb 9;22(1):67. doi: 10.1186/s12935-022-02485-2. Cancer Cell Int. 2022. PMID: 35139863 Free PMC article.
-
Identification of PDZD11 as a Potential Biomarker Associated with Immune Infiltration for Diagnosis and Prognosis in Epithelial Ovarian Cancer.Int J Gen Med. 2024 May 14;17:2113-2128. doi: 10.2147/IJGM.S459418. eCollection 2024. Int J Gen Med. 2024. PMID: 38766598 Free PMC article.
-
PLEKHA7 Recruits PDZD11 to Adherens Junctions to Stabilize Nectins.J Biol Chem. 2016 May 20;291(21):11016-29. doi: 10.1074/jbc.M115.712935. Epub 2016 Apr 4. J Biol Chem. 2016. PMID: 27044745 Free PMC article.
Cited by
-
PPIH acts as a potential predictive biomarker for patients with common solid tumors.BMC Cancer. 2024 Jun 4;24(1):681. doi: 10.1186/s12885-024-12446-9. BMC Cancer. 2024. PMID: 38834966 Free PMC article.
-
Long non-coding RNAs in ferroptosis and cuproptosis impact on prognosis and treatment in hepatocellular carcinoma.Clin Exp Med. 2024 Jun 22;24(1):135. doi: 10.1007/s10238-024-01397-x. Clin Exp Med. 2024. PMID: 38907744 Free PMC article.
-
Proto-Oncogene FAM50A Can Regulate the Immune Microenvironment and Development of Hepatocellular Carcinoma In Vitro and In Vivo.Int J Mol Sci. 2023 Feb 6;24(4):3217. doi: 10.3390/ijms24043217. Int J Mol Sci. 2023. PMID: 36834630 Free PMC article.
-
Evaluation of PDZD11 in hepatocellular carcinoma: prognostic value and diagnostic potential in combination with AFP.Front Oncol. 2025 Mar 25;15:1533865. doi: 10.3389/fonc.2025.1533865. eCollection 2025. Front Oncol. 2025. PMID: 40201341 Free PMC article.
-
PPIH gene regulation system and its prognostic significance in hepatocellular carcinoma: a comprehensive analysis.Aging (Albany NY). 2023 Oct 23;15(20):11448-11470. doi: 10.18632/aging.205134. Epub 2023 Oct 23. Aging (Albany NY). 2023. PMID: 37874737 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

