Genome-Wide Detection of Copy Number Variations and Their Association With Distinct Phenotypes in the World's Sheep
- PMID: 34093663
- PMCID: PMC8175073
- DOI: 10.3389/fgene.2021.670582
Genome-Wide Detection of Copy Number Variations and Their Association With Distinct Phenotypes in the World's Sheep
Abstract
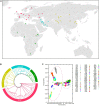
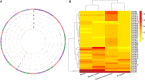
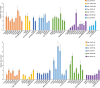
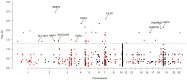
Copy number variations (CNVs) are a major source of structural variation in mammalian genomes. Here, we characterized the genome-wide CNV in 2059 sheep from 67 populations all over the world using the Ovine Infinium HD (600K) SNP BeadChip. We tested their associations with distinct phenotypic traits by conducting multiple independent genome-wide tests. In total, we detected 7547 unique CNVs and 18,152 CNV events in 1217 non-redundant CNV regions (CNVRs), covering 245 Mb (∼10%) of the whole sheep genome. We identified seven CNVRs with frequencies correlating to geographical origins and 107 CNVRs overlapping 53 known quantitative trait loci (QTLs). Gene ontology and pathway enrichment analyses of CNV-overlapping genes revealed their common involvement in energy metabolism, endocrine regulation, nervous system development, cell proliferation, immune, and reproduction. For the phenotypic traits, we detected significantly associated (adjusted P < 0.05) CNVRs harboring functional candidate genes, such as SBNO2 for polycerate; PPP1R11 and GABBR1 for tail weight; AKT1 for supernumerary nipple; CSRP1, WNT7B, HMX1, and FGFR3 for ear size; and NOS3 and FILIP1 in Wadi sheep; SNRPD3, KHDRBS2, and SDCCAG3 in Hu sheep; NOS3, BMP1, and SLC19A1 in Icelandic; CDK2 in Finnsheep; MICA in Romanov; and REEP4 in Texel sheep for litter size. These CNVs and associated genes are important markers for molecular breeding of sheep and other livestock species.
Keywords: CNVs; GWAS; genetic adaptation; selection; sheep.
Copyright © 2021 Salehian-Dehkordi, Xu, Xu, Li, Luo, Liu, Wang, Cao, Shen, Gao, Chen, Glessner, Lenstra, Esmailizadeh, Li and Lv.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
-
Identification of copy number variation in Tibetan sheep using whole genome resequencing reveals evidence of genomic selection.BMC Genomics. 2023 Sep 19;24(1):555. doi: 10.1186/s12864-023-09672-z. BMC Genomics. 2023. PMID: 37726692 Free PMC article.
-
Analysis of copy number variations in the sheep genome using 50K SNP BeadChip array.BMC Genomics. 2013 Apr 8;14:229. doi: 10.1186/1471-2164-14-229. BMC Genomics. 2013. PMID: 23565757 Free PMC article.
-
Genome-wide detection of copy number variation in Chinese indigenous sheep using an ovine high-density 600 K SNP array.Sci Rep. 2017 Apr 19;7(1):912. doi: 10.1038/s41598-017-00847-9. Sci Rep. 2017. PMID: 28424525 Free PMC article.
-
The challenges and importance of structural variation detection in livestock.Front Genet. 2014 Feb 18;5:37. doi: 10.3389/fgene.2014.00037. eCollection 2014. Front Genet. 2014. PMID: 24600474 Free PMC article. Review.
Cited by
-
Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness.Animals (Basel). 2023 Feb 15;13(4):674. doi: 10.3390/ani13040674. Animals (Basel). 2023. PMID: 36830461 Free PMC article.
-
Identification of Copy Number Variations and Genetic Diversity in Italian Insular Sheep Breeds.Animals (Basel). 2022 Jan 17;12(2):217. doi: 10.3390/ani12020217. Animals (Basel). 2022. PMID: 35049839 Free PMC article.
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
-
Genomic Landscape of Copy Number Variations and Their Associations with Climatic Variables in the World's Sheep.Genes (Basel). 2023 Jun 13;14(6):1256. doi: 10.3390/genes14061256. Genes (Basel). 2023. PMID: 37372436 Free PMC article.
-
Genome-wide detection of copy number variation in American mink using whole-genome sequencing.BMC Genomics. 2022 Sep 13;23(1):649. doi: 10.1186/s12864-022-08874-1. BMC Genomics. 2022. PMID: 36096727 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

