Circ-MFN2 Positively Regulates the Proliferation, Metastasis, and Radioresistance of Colorectal Cancer by Regulating the miR-574-3p/IGF1R Signaling Axis
- PMID: 34093664
- PMCID: PMC8170135
- DOI: 10.3389/fgene.2021.671337
Circ-MFN2 Positively Regulates the Proliferation, Metastasis, and Radioresistance of Colorectal Cancer by Regulating the miR-574-3p/IGF1R Signaling Axis
Abstract
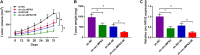
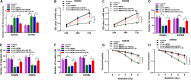
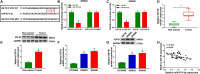
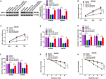
Numerous studies have shown that the expression of circular RNA (circRNA) is closely related to the malignant progression of cancer. However, the role of circ-MFN2 in colorectal cancer (CRC) is unclear. Our study aims to explore the role and mechanism of circ-MFN2 in CRC progression. The relative expression levels of circ-MFN2, microRNA (miR)-574-3p and insulin-like growth factor 1 receptor (IGF1R) were detected by quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability was determined using 3-(4, 5-dimethyl-2 thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay. The colony number and radioresistance of cells were assessed using colony formation assay. Moreover, the migration and invasion of cells were measured using transwell assay. Tumor xenograft model was constructed to evaluate the effect of circ-MFN2 knockdown on CRC tumor growth. Furthermore, dual-luciferase reporter assay was used to verify the interaction between miR-574-3p and circ-MFN2 or IGF1R. In addition, the protein level of IGF1R was evaluated by western blot (WB) analysis. Circ-MFN2 expression was elevated in CRC tissues and cells. Knockdown of circ-MFN2 restrained the proliferation, migration, invasion, and radioresistance of CRC cells in vitro. Furthermore, silenced circ-MFN2 also reduced the tumor volume and weight of CRC in vivo. MiR-574-3p could be sponged by circ-MFN2, and its inhibitor reversed the suppression effect of circ-MFN2 silencing on CRC progression. Moreover, IGF1R was a target of miR-574-3p, and its overexpression reversed the inhibition effect of miR-574-3p mimic on CRC progression. In addition, circ-MFN2 could positively regulate IGF1R expression by sponging miR-574-3p. Our results revealed that circ-MFN2 promoted the proliferation, metastasis and radioresistance of CRC through regulating the miR-574-3p/IGF1R axis, suggesting that circ-MFN2 might be a novel therapeutic biomarker for CRC.
Keywords: CRC; IGF1R; circ-MFN2; miR-574-3p; progression.
Copyright © 2021 Liu, Peng, Li and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis.Eur J Clin Invest. 2021 Dec;51(12):e13607. doi: 10.1111/eci.13607. Epub 2021 Jun 4. Eur J Clin Invest. 2021. PMID: 34085707
-
Circ_0006174 Upregulates IGF1R to Enhance Radioresistance and Tumorigenesis in Colorectal Cancer via miR-940 Suppression.Appl Biochem Biotechnol. 2025 Jan;197(1):497-517. doi: 10.1007/s12010-024-05028-9. Epub 2024 Aug 22. Appl Biochem Biotechnol. 2025. PMID: 39172343
-
Circ_0011385 knockdown inhibits cell proliferation, migration and invasion, whereas promotes cell apoptosis by regulating miR-330-3p/MYO6 axis in colorectal cancer.Biomed J. 2023 Feb;46(1):110-121. doi: 10.1016/j.bj.2022.01.007. Epub 2022 Jan 26. Biomed J. 2023. PMID: 35091088 Free PMC article.
-
Circ_0067835 Knockdown Enhances the Radiosensitivity of Colorectal Cancer by miR-296-5p/IGF1R Axis.Onco Targets Ther. 2021 Jan 18;14:491-502. doi: 10.2147/OTT.S281011. eCollection 2021. Onco Targets Ther. 2021. PMID: 33500625 Free PMC article.
-
Circ_0084188 promotes colorectal cancer progression by sponging miR-654-3p and regulating kruppel-like factor 12.Kaohsiung J Med Sci. 2023 Nov;39(11):1062-1076. doi: 10.1002/kjm2.12749. Epub 2023 Sep 12. Kaohsiung J Med Sci. 2023. PMID: 37698263 Free PMC article.
Cited by
-
Circular RNAs modulate Hippo-YAP signaling: functional mechanisms in cancer.Theranostics. 2022 May 16;12(9):4269-4287. doi: 10.7150/thno.71708. eCollection 2022. Theranostics. 2022. PMID: 35673576 Free PMC article. Review.
-
Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages.Bioengineered. 2022 Mar;13(3):6243-6256. doi: 10.1080/21655979.2021.2003929. Bioengineered. 2022. PMID: 35349390 Free PMC article.
-
Role of non-coding RNAs in radiosensitivity of colorectal cancer: A narrative review.Front Oncol. 2022 Jul 22;12:889658. doi: 10.3389/fonc.2022.889658. eCollection 2022. Front Oncol. 2022. PMID: 35936676 Free PMC article. Review.
-
CircRNAs in Tumor Radioresistance.Biomolecules. 2022 Oct 28;12(11):1586. doi: 10.3390/biom12111586. Biomolecules. 2022. PMID: 36358938 Free PMC article. Review.
-
Biological functions, mechanisms, and clinical significance of circular RNA in colorectal cancer.Front Oncol. 2023 Mar 6;13:1138481. doi: 10.3389/fonc.2023.1138481. eCollection 2023. Front Oncol. 2023. PMID: 36950552 Free PMC article. Review.
References
-
- Chitnis M. M., Yuen J. S., Protheroe A. S., Pollak M., Macaulay V. M. (2008). The type 1 insulin-like growth factor receptor pathway. Clin. Cancer Res. 14 6364–6370. - PubMed
LinkOut - more resources
Full Text Sources

