Towards multi-label classification: Next step of machine learning for microbiome research
- PMID: 34093989
- PMCID: PMC8131981
- DOI: 10.1016/j.csbj.2021.04.054
Towards multi-label classification: Next step of machine learning for microbiome research
Abstract
Machine learning (ML) has been widely used in microbiome research for biomarker selection and disease prediction. By training microbial profiles of samples from patients and healthy controls, ML classifiers constructs data models by community features that highly correlated with the target diseases, so as to determine the status of new samples. To clearly understand the host-microbe interaction of specific diseases, previous studies always focused on well-designed cohorts, in which each sample was exactly labeled by a single status type. However, in fact an individual may be associated with multiple diseases simultaneously, which introduce additional variations on microbial patterns that interferes the status detection. More importantly, comorbidities or complications can be missed by regular ML models, limiting the practical application of microbiome techniques. In this review, we summarize the typical ML approaches of single-label classification for microbiome research, and demonstrate their limitations in multi-label disease detection using a real dataset. Then we prospect a further step of ML towards multi-label classification that potentially solves the aforementioned problem, including a series of promising strategies and key technical issues for applying multi-label classification in microbiome-based studies.
Keywords: Machine learning; Microbiome; Multi-label classification; Single-label classification.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
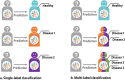
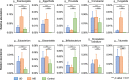


Similar articles
-
Co-Labeling for Multi-View Weakly Labeled Learning.IEEE Trans Pattern Anal Mach Intell. 2016 Jun;38(6):1113-25. doi: 10.1109/TPAMI.2015.2476813. Epub 2015 Sep 4. IEEE Trans Pattern Anal Mach Intell. 2016. PMID: 26353366
-
Gene-based microbiome representation enhances host phenotype classification.mSystems. 2023 Aug 31;8(4):e0053123. doi: 10.1128/msystems.00531-23. Epub 2023 Jul 5. mSystems. 2023. PMID: 37404032 Free PMC article.
-
The future of Cochrane Neonatal.Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834
-
Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment.Front Microbiol. 2021 Feb 19;12:634511. doi: 10.3389/fmicb.2021.634511. eCollection 2021. Front Microbiol. 2021. PMID: 33737920 Free PMC article. Review.
-
Machine learning for data integration in human gut microbiome.Microb Cell Fact. 2022 Nov 23;21(1):241. doi: 10.1186/s12934-022-01973-4. Microb Cell Fact. 2022. PMID: 36419034 Free PMC article. Review.
Cited by
-
Parallel-Meta Suite: Interactive and rapid microbiome data analysis on multiple platforms.Imeta. 2022 Mar 6;1(1):e1. doi: 10.1002/imt2.1. eCollection 2022 Mar. Imeta. 2022. PMID: 38867729 Free PMC article.
-
An Improved Diagnostic of the Mycobacterium tuberculosis Drug Resistance Status by Applying a Decision Tree to Probabilities Assigned by the CatBoost Multiclassifier of Matrix Metalloproteinases Biomarkers.Diagnostics (Basel). 2022 Nov 17;12(11):2847. doi: 10.3390/diagnostics12112847. Diagnostics (Basel). 2022. PMID: 36428907 Free PMC article.
-
PhyloMix: enhancing microbiome-trait association prediction through phylogeny-mixing augmentation.Bioinformatics. 2025 Feb 4;41(2):btaf014. doi: 10.1093/bioinformatics/btaf014. Bioinformatics. 2025. PMID: 39799515 Free PMC article.
-
Comprehensive Assessment of 16S rRNA Gene Amplicon Sequencing for Microbiome Profiling across Multiple Habitats.Microbiol Spectr. 2023 Jun 15;11(3):e0056323. doi: 10.1128/spectrum.00563-23. Epub 2023 Apr 27. Microbiol Spectr. 2023. PMID: 37102867 Free PMC article.
-
Advanced computational tools, artificial intelligence and machine-learning approaches in gut microbiota and biomarker identification.Front Med Technol. 2025 Apr 15;6:1434799. doi: 10.3389/fmedt.2024.1434799. eCollection 2024. Front Med Technol. 2025. PMID: 40303946 Free PMC article. Review.
References
-
- Knight R. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. - PubMed
-
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. - PubMed
-
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
