Investigation of cellular uptake mechanism of functionalised gold nanoparticles into breast cancer using SERS
- PMID: 34094083
- PMCID: PMC8159335
- DOI: 10.1039/d0sc01255f
Investigation of cellular uptake mechanism of functionalised gold nanoparticles into breast cancer using SERS
Abstract
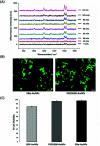
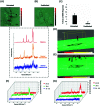
Gold nanoparticles (AuNPs) are widely used in various applications such as cancer imaging and drug delivery. The functionalisation of AuNPs has been shown to affect their cellular internalisation, accumulation and targeting efficiency. The mechanism of cellular uptake of functionalised AuNPs by different cancer cells is not well understood. Therefore, a detailed understanding of the molecular processes is necessary to improve AuNPs for their selective uptake and fate in specific cellular systems. This knowledge can greatly help in designing nanotags with higher cellular uptake for more selective and specific targeting capabilities with less off-target effects. Here, we demonstrate for the first time a straightforward and non-destructive 3D surface enhanced Raman spectroscopy (SERS) imaging approach to track the cellular uptake and localisation of AuNPs functionalised with an anti-ERα (estrogen receptor alpha) antibody in MCF-7 ERα-positive human breast cancer cells under different conditions including temperature and dynamin inhibition. 3D SERS enabled information rich monitoring of the intracellular internalisation of the SERS nanotags. It was found that ERα-AuNPs were internalised by MCF-7 cells in a temperature-dependent manner suggesting an active endocytosis-dependent mechanism. 3D SERS cell mapping also indicated that the nanotags entered MCF-7 cells using dynamin dependent endocytosis, since dynamin inhibition resulted in the SERS signal being obtained from, or close to, the cell surface rather than inside the cells. Finally, ERα-AuNPs were found to enter MCF-7 cells using an ERα receptor-mediated endocytosis process. This study addresses the role of functionalisation of SERS nanotags in biological environments and highlights the benefits of using 3D SERS for the investigation of cellular uptake processes.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Characterisation of estrogen receptor alpha (ERα) expression in breast cancer cells and effect of drug treatment using targeted nanoparticles and SERS.Analyst. 2020 Nov 9;145(22):7225-7233. doi: 10.1039/d0an01532f. Analyst. 2020. PMID: 33164013
-
Detection of Estrogen Receptor Alpha and Assessment of Fulvestrant Activity in MCF-7 Tumor Spheroids Using Microfluidics and SERS.Anal Chem. 2021 Apr 13;93(14):5862-5871. doi: 10.1021/acs.analchem.1c00188. Epub 2021 Apr 2. Anal Chem. 2021. PMID: 33797884 Free PMC article.
-
Rhodamine 6G conjugated to gold nanoparticles as labels for both SERS and fluorescence studies on live endothelial cells.Mikrochim Acta. 2015;182(1):119-127. doi: 10.1007/s00604-014-1307-5. Epub 2014 Jun 19. Mikrochim Acta. 2015. PMID: 25568498 Free PMC article.
-
Gold Fluorescence Nanoparticles for Enhanced SERS Detection in Biomedical Sensor Applications: Current Trends and Future Directions.Chem Rec. 2024 Feb 5:e202300303. doi: 10.1002/tcr.202300303. Online ahead of print. Chem Rec. 2024. PMID: 38314935 Review.
-
Designing SERS nanotags for profiling overexpressed surface markers on single cancer cells: A review.Talanta. 2024 Aug 15;276:126225. doi: 10.1016/j.talanta.2024.126225. Epub 2024 May 9. Talanta. 2024. PMID: 38749157 Review.
Cited by
-
Designing Functional Bionanoconstructs for Effective In Vivo Targeting.Bioconjug Chem. 2022 Mar 16;33(3):429-443. doi: 10.1021/acs.bioconjchem.1c00546. Epub 2022 Feb 15. Bioconjug Chem. 2022. PMID: 35167255 Free PMC article. Review.
-
New Model for Quantifying the Nanoparticle Concentration Using SERS Supported by Multimodal Mass Spectrometry.Anal Chem. 2023 Feb 7;95(5):2757-2764. doi: 10.1021/acs.analchem.2c03779. Epub 2023 Jan 26. Anal Chem. 2023. PMID: 36701560 Free PMC article.
-
Intelligent Bio-Responsive Fluorescent Au-shRNA Complexes for Regulated Autophagy and Effective Cancer Bioimaging and Therapeutics.Biosensors (Basel). 2021 Oct 28;11(11):425. doi: 10.3390/bios11110425. Biosensors (Basel). 2021. PMID: 34821640 Free PMC article.
-
Enhancing the therapeutic efficacy of nanoparticles for cancer treatment using versatile targeted strategies.J Hematol Oncol. 2022 Sep 12;15(1):132. doi: 10.1186/s13045-022-01320-5. J Hematol Oncol. 2022. PMID: 36096856 Free PMC article. Review.
-
Application of Nanohybrid Substrates with Layer-by-Layer Self-Assembling Properties to High-Sensitivity Surface-Enhanced Raman Scattering Detection.ACS Omega. 2023 Dec 26;9(1):1894-1903. doi: 10.1021/acsomega.3c08608. eCollection 2024 Jan 9. ACS Omega. 2023. PMID: 38222643 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

