Melting temperature measurement and mesoscopic evaluation of single, double and triple DNA mismatches
- PMID: 34094181
- PMCID: PMC8163305
- DOI: 10.1039/d0sc01700k
Melting temperature measurement and mesoscopic evaluation of single, double and triple DNA mismatches
Abstract
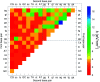
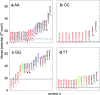
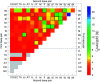
Unlike the canonical base pairs AT and GC, the molecular properties of mismatches such as hydrogen bonding and stacking interactions are strongly dependent on the identity of the neighbouring base pairs. As a result, due to the sheer number of possible combinations of mismatches and flanking base pairs, only a fraction of these have been studied in varying experiments or theoretical models. Here, we report on the melting temperature measurement and mesoscopic analysis of contiguous DNA mismatches in nearest-neighbours and next-nearest neighbour contexts. A total of 4032 different mismatch combinations, including single, double and triple mismatches were covered. These were compared with 64 sequences containing all combinations of canonical base pairs in the same location under the same conditions. For a substantial number of single mismatch configurations, 15%, the measured melting temperatures were higher than the least stable AT base pair. The mesoscopic calculation, using the Peyrard-Bishop model, was performed on the set of 4096 sequences, and resulted in estimates of on-site and nearest-neighbour interactions that can be correlated to hydrogen bonding and base stacking. Our results confirm many of the known properties of mismatches, including the peculiar sheared stacking of tandem GA mismatches. More intriguingly, it also reveals that a number of mismatches present strong hydrogen bonding when flanked on both sites by other mismatches. To highlight the applicability of our results, we discuss a number of practical situations such as enzyme binding affinities, thymine DNA glycosylase repair activity, and trinucleotide repeat expansions.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures







Similar articles
-
Evaluating Hydrogen Bonds and Base Stacking of Single, Tandem and Terminal GU Mismatches in RNA with a Mesoscopic Model.J Chem Inf Model. 2016 Jan 25;56(1):101-9. doi: 10.1021/acs.jcim.5b00571. Epub 2015 Dec 17. J Chem Inf Model. 2016. PMID: 26624232
-
The thermal stability of DNA fragments with tandem mismatches at a d(CXYG).d(CY'X'G) site.Nucleic Acids Res. 1996 Feb 15;24(4):707-12. doi: 10.1093/nar/24.4.707. Nucleic Acids Res. 1996. PMID: 8604314 Free PMC article.
-
Thermodynamics of DNA duplexes with adjacent G.A mismatches.Biochemistry. 1991 Jul 30;30(30):7566-72. doi: 10.1021/bi00244a028. Biochemistry. 1991. PMID: 1854755
-
Influence of pH on the conformation and stability of mismatch base-pairs in DNA.J Mol Biol. 1990 Apr 5;212(3):437-40. doi: 10.1016/0022-2836(90)90320-L. J Mol Biol. 1990. PMID: 2182883 Review.
-
Unusual DNA duplex and hairpin motifs.Nucleic Acids Res. 2003 May 15;31(10):2461-74. doi: 10.1093/nar/gkg367. Nucleic Acids Res. 2003. PMID: 12736295 Free PMC article. Review.
Cited by
-
Nearest-neighbour transition-state analysis for nucleic acid kinetics.Nucleic Acids Res. 2021 May 7;49(8):4574-4585. doi: 10.1093/nar/gkab205. Nucleic Acids Res. 2021. PMID: 33823552 Free PMC article.
-
A pH and Mg2+-Responsive Molecular Switch Based on a Stable DNA Minidumbbell Bearing 5' and 3'-Overhangs.ACS Omega. 2021 Oct 14;6(42):28263-28269. doi: 10.1021/acsomega.1c04346. eCollection 2021 Oct 26. ACS Omega. 2021. PMID: 34723023 Free PMC article.
-
Re-pairing DNA: binding of a ruthenium phi complex to a double mismatch.Chem Sci. 2024 May 16;15(24):9096-9103. doi: 10.1039/d4sc01448k. eCollection 2024 Jun 19. Chem Sci. 2024. PMID: 38903237 Free PMC article.
-
Non-linear Hamiltonian models for DNA.Eur Biophys J. 2022 Sep;51(6):431-447. doi: 10.1007/s00249-022-01614-z. Epub 2022 Aug 17. Eur Biophys J. 2022. PMID: 35976412 Review.
-
Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.Nucleic Acids Res. 2022 Aug 26;50(15):8867-8881. doi: 10.1093/nar/gkac629. Nucleic Acids Res. 2022. PMID: 35871296 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

