Characterization of Plasma Cell-Free DNA Integrity Using Droplet-Based Digital PCR: Toward the Development of Circulating Tumor DNA-Dedicated Assays
- PMID: 34094923
- PMCID: PMC8174096
- DOI: 10.3389/fonc.2021.639675
Characterization of Plasma Cell-Free DNA Integrity Using Droplet-Based Digital PCR: Toward the Development of Circulating Tumor DNA-Dedicated Assays
Abstract
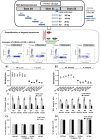
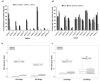
Background: Cellular-cell free-DNA (ccfDNA) is being explored as a diagnostic and prognostic tool for various diseases including cancer. Beyond the evaluation of the ccfDNA mutational status, its fragmentation has been investigated as a potential cancer biomarker in several studies. However, probably due to a lack of standardized procedures dedicated to preanalytical and analytical processing of plasma samples, contradictory results have been published. Methods: ddPCR assays allowing the detection of KRAS wild-type and mutated sequences (KRAS p.G12V, pG12D, and pG13D) were designed to target different fragments sizes. Once validated on fragmented and non-fragmented DNA extracted from cancer cell lines, these assays were used to investigate the influence of the extraction methods on the non-mutated and mutated ccfDNA integrity reflected by the DNA integrity index (DII). The DII was then analyzed in two prospective cohorts of metastatic colorectal cancer patients (RASANC study n = 34; PLACOL study n = 12) and healthy subjects (n = 49). Results and Discussion: Our results demonstrate that ccfDNA is highly fragmented in mCRC patients compared with healthy individuals. These results strongly suggest that the characterization of ccfDNA integrity hold great promise toward the development of a universal biomarker for the follow-up of mCRC patients. Furthermore, they support the importance of standardization of sample handling and processing in such analysis.
Keywords: DNA integrity index; apoptosis; cancer biomarker; circulating cell-free DNA; circulating tumor DNA; necrosis; picoliter-droplet digital PCR.
Copyright © 2021 Poulet, Garlan, Garrigou, Zonta, Benhaim, Carrillon, Didelot, Le Corre, Mulot, Nizard, Ginot, Boutonnet-Rodat, Blons, Bachet, Taïeb, Zaanan, Geromel, Pellegrina, Laurent-Puig, Wang-Renault and Taly.
Conflict of interest statement
VT: Honoraria from Raindance Technologies and Boerhinger Ingehleim; cofounder emulseo; board emulseo. JT: Honoraria from Merck, Amgen, Roche, Pierre Fabre, MSD, Sanofi, and Lilly, Servier, and Astra-Zeneca. AZ: consulting and/or advisory boards for: Roche, Merck Serono, Amgen, Sanofi, and Lilly. PL-P: Honoraria and board: Amgen, Merck-Serono, Boehringer Ingelheim, Sanofi, Roche, and Lilly. HB: Honoraria Astra-Zeneca, BMS, MSD. JT: Honoraria from Merck, AMGEN, ROCHE, SIRTEX, BAXALTA, SANOFI, Lilly, and Servier. AZ: Consulting and/or advisory boards for Roche, Merck Serono, Amgen, Sanofi, and Lilly. GP, LP, and VG: Eurofins-Biomnis employee. FGa: Bio-Rad employee. AB and FGi were employed by company ADELIS. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows.Clin Chem. 2020 Jan 1;66(1):149-160. doi: 10.1373/clinchem.2019.306837. Clin Chem. 2020. PMID: 31628139
-
KRAS G12V Mutation Detection by Droplet Digital PCR in Circulating Cell-Free DNA of Colorectal Cancer Patients.Int J Mol Sci. 2016 Apr 1;17(4):484. doi: 10.3390/ijms17040484. Int J Mol Sci. 2016. PMID: 27043547 Free PMC article.
-
Circulating cell free DNA: Preanalytical considerations.Clin Chim Acta. 2013 Sep 23;424:222-30. doi: 10.1016/j.cca.2013.05.022. Epub 2013 May 30. Clin Chim Acta. 2013. PMID: 23727028 Review.
-
Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer.Mol Oncol. 2014 Jul;8(5):927-41. doi: 10.1016/j.molonc.2014.02.005. Epub 2014 Mar 24. Mol Oncol. 2014. PMID: 24698732 Free PMC article.
-
Circulating Plasma Tumor DNA.Adv Exp Med Biol. 2016;882:259-76. doi: 10.1007/978-3-319-22909-6_11. Adv Exp Med Biol. 2016. PMID: 26987539 Review.
Cited by
-
An Updated Review on Molecular Biomarkers in Diagnosis and Therapy of Colorectal Cancer.Curr Cancer Drug Targets. 2024;24(6):595-611. doi: 10.2174/0115680096270555231113074003. Curr Cancer Drug Targets. 2024. PMID: 38031267 Review.
-
The utility of cfDNA in TGCT patient management: a systematic review.Ther Adv Med Oncol. 2022 May 25;14:17588359221090365. doi: 10.1177/17588359221090365. eCollection 2022. Ther Adv Med Oncol. 2022. PMID: 35656387 Free PMC article. Review.
-
Circadian rhythm and circulating cell-free DNA release on healthy subjects.Sci Rep. 2023 Dec 7;13(1):21675. doi: 10.1038/s41598-023-47851-w. Sci Rep. 2023. PMID: 38065990 Free PMC article.
References
-
- Panabières CA, Pantel K. Clinical applications of circulating tumor cells and circulating tumor dna as liquid biopsy. Cancer Discovery. (2016) 6:479–91. 10.1158/2159-8290.CD-15-1483 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

