Salmonella Biofilms Tolerate Hydrogen Peroxide by a Combination of Extracellular Polymeric Substance Barrier Function and Catalase Enzymes
- PMID: 34095002
- PMCID: PMC8171120
- DOI: 10.3389/fcimb.2021.683081
Salmonella Biofilms Tolerate Hydrogen Peroxide by a Combination of Extracellular Polymeric Substance Barrier Function and Catalase Enzymes
Abstract
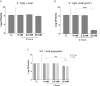
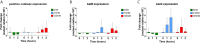
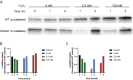
The ability of Salmonella enterica subspecies enterica serovar Typhi (S. Typhi) to cause chronic gallbladder infections is dependent on biofilm growth on cholesterol gallstones. Non-typhoidal Salmonella (e.g. S. Typhimurium) also utilize the biofilm state to persist in the host and the environment. How the pathogen maintains recalcitrance to the host response, and oxidative stress in particular, during chronic infection is poorly understood. Previous experiments demonstrated that S. Typhi and S. Typhimurium biofilms are tolerant to hydrogen peroxide (H2O2), but that mutations in the biofilm extracellular polymeric substances (EPSs) O antigen capsule, colanic acid, or Vi antigen reduce tolerance. Here, biofilm-mediated tolerance to oxidative stress was investigated using a combination of EPS and catalase mutants, as catalases are important detoxifiers of H2O2. Using co-cultured biofilms of wild-type (WT) bacteria with EPS mutants, it was demonstrated that colanic acid in S. Typhimurium and Vi antigen in S. Typhi have a community function and protect all biofilm-resident bacteria rather than to only protect the individual cells producing the EPSs. However, the H2O2 tolerance deficiency of a O antigen capsule mutant was unable to be compensated for by co-culture with WT bacteria. For curli fimbriae, both WT and mutant strains are tolerant to H2O2 though unexpectedly, co-cultured WT/mutant biofilms challenged with H2O2 resulted in sensitization of both strains, suggesting a more nuanced oxidative resistance alteration in these co-cultures. Three catalase mutant (katE, katG and a putative catalase) biofilms were also examined, demonstrating significant reductions in biofilm H2O2 tolerance for the katE and katG mutants. Biofilm co-culture experiments demonstrated that catalases exhibit a community function. We further hypothesized that biofilms are tolerant to H2O2 because the physical barrier formed by EPSs slows penetration of H2O2 into the biofilm to a rate that can be mitigated by intra-biofilm catalases. Compared to WT, EPS-deficient biofilms have a heighted response even to low-dose (2.5 mM) H2O2 challenge, confirming that resident bacteria of EPS-deficient biofilms are under greater stress and have limited protection from H2O2. Thus, these data provide an explanation for how Salmonella achieves tolerance to H2O2 by a combination of an EPS-mediated barrier and enzymatic detoxification.
Keywords: Salmonella; biofilms; chronic infection; extracellular polymeric substances (EPSs); hydrogen peroxide; innate immunity.
Copyright © 2021 Hahn, González and Gunn.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


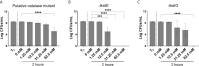

References
-
- Barthel M., Hapfelmeier S., Quintanilla-Martínez L., Kremer M., Rohde M., Hogardt M., et al. (2003). Pretreatment of Mice With Streptomycin Provides a Salmonella Enterica Serovar Typhimurium Colitis Model That Allows Analysis of Both Pathogen and Host. Infect. Immun. 71 (5), 2839–2858. 10.1128/IAI.71.5.2839-2858.2003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

