Delayed Rise of Oral Fluid Antibodies, Elevated BMI, and Absence of Early Fever Correlate With Longer Time to SARS-CoV-2 RNA Clearance in a Longitudinally Sampled Cohort of COVID-19 Outpatients
- PMID: 34095338
- PMCID: PMC8083254
- DOI: 10.1093/ofid/ofab195
Delayed Rise of Oral Fluid Antibodies, Elevated BMI, and Absence of Early Fever Correlate With Longer Time to SARS-CoV-2 RNA Clearance in a Longitudinally Sampled Cohort of COVID-19 Outpatients
Abstract
Background: Sustained molecular detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in the upper respiratory tract (URT) in mild to moderate coronavirus disease 2019 (COVID-19) is common. We sought to identify host and immune determinants of prolonged SARS-CoV-2 RNA detection.
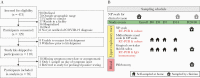
Methods: Ninety-five symptomatic outpatients self-collected midturbinate nasal, oropharyngeal (OP), and gingival crevicular fluid (oral fluid) samples at home and in a research clinic a median of 6 times over 1-3 months. Samples were tested for viral RNA, virus culture, and SARS-CoV-2 and other human coronavirus antibodies, and associations were estimated using Cox proportional hazards models.
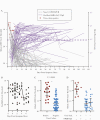
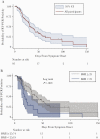
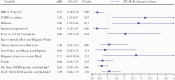
Results: Viral RNA clearance, as measured by SARS-CoV-2 reverse transcription polymerase chain reaction (RT-PCR), in 507 URT samples occurred a median (interquartile range) 33.5 (17-63.5) days post-symptom onset. Sixteen nasal-OP samples collected 2-11 days post-symptom onset were virus culture positive out of 183 RT-PCR-positive samples tested. All participants but 1 with positive virus culture were negative for concomitant oral fluid anti-SARS-CoV-2 antibodies. The mean time to first antibody detection in oral fluid was 8-13 days post-symptom onset. A longer time to first detection of oral fluid anti-SARS-CoV-2 S antibodies (adjusted hazard ratio [aHR], 0.96; 95% CI, 0.92-0.99; P = .020) and body mass index (BMI) ≥25 kg/m2 (aHR, 0.37; 95% CI, 0.18-0.78; P = .009) were independently associated with a longer time to SARS-CoV-2 viral RNA clearance. Fever as 1 of first 3 COVID-19 symptoms correlated with shorter time to viral RNA clearance (aHR, 2.06; 95% CI, 1.02-4.18; P = .044).
Conclusions: We demonstrate that delayed rise of oral fluid SARS-CoV-2-specific antibodies, elevated BMI, and absence of early fever are independently associated with delayed URT viral RNA clearance.
Keywords: COVID-19; RT-PCR; SARS-CoV-2; antibody; viral RNA.
© The Author(s) 2021. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Figures

Update of
-
Delayed rise of oral fluid antibodies, elevated BMI, and absence of early fever correlate with longer time to SARS-CoV-2 RNA clearance in an longitudinally sampled cohort of COVID-19 outpatients.medRxiv [Preprint]. 2021 Mar 3:2021.03.02.21252420. doi: 10.1101/2021.03.02.21252420. medRxiv. 2021. Update in: Open Forum Infect Dis. 2021 Apr 16;8(6):ofab195. doi: 10.1093/ofid/ofab195. PMID: 33688688 Free PMC article. Updated. Preprint.
Similar articles
-
Delayed rise of oral fluid antibodies, elevated BMI, and absence of early fever correlate with longer time to SARS-CoV-2 RNA clearance in an longitudinally sampled cohort of COVID-19 outpatients.medRxiv [Preprint]. 2021 Mar 3:2021.03.02.21252420. doi: 10.1101/2021.03.02.21252420. medRxiv. 2021. Update in: Open Forum Infect Dis. 2021 Apr 16;8(6):ofab195. doi: 10.1093/ofid/ofab195. PMID: 33688688 Free PMC article. Updated. Preprint.
-
Infectious Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Virus in Symptomatic Coronavirus Disease 2019 (COVID-19) Outpatients: Host, Disease, and Viral Correlates.Clin Infect Dis. 2022 Aug 24;75(1):e1028-e1036. doi: 10.1093/cid/ciab968. Clin Infect Dis. 2022. PMID: 35022711 Free PMC article.
-
Long COVID brain fog and muscle pain are associated with longer time to clearance of SARS-CoV-2 RNA from the upper respiratory tract during acute infection.Front Immunol. 2023 Apr 28;14:1147549. doi: 10.3389/fimmu.2023.1147549. eCollection 2023. Front Immunol. 2023. PMID: 37187756 Free PMC article.
-
At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data.BMC Med. 2020 Nov 4;18(1):346. doi: 10.1186/s12916-020-01810-8. BMC Med. 2020. PMID: 33143712 Free PMC article.
-
[SARS-CoV-2 and Microbiological Diagnostic Dynamics in COVID-19 Pandemic].Mikrobiyol Bul. 2020 Jul;54(3):497-509. doi: 10.5578/mb.69839. Mikrobiyol Bul. 2020. PMID: 32755524 Review. Turkish.
Cited by
-
Methodological approaches to optimize multiplex oral fluid SARS-CoV-2 IgG assay performance and correlation with serologic and neutralizing antibody responses.J Immunol Methods. 2023 Mar;514:113440. doi: 10.1016/j.jim.2023.113440. Epub 2023 Feb 10. J Immunol Methods. 2023. PMID: 36773929 Free PMC article.
-
Application of machine learning algorithms to identify serological predictors of COVID-19 severity and outcomes.Commun Med (Lond). 2024 Nov 26;4(1):249. doi: 10.1038/s43856-024-00658-w. Commun Med (Lond). 2024. PMID: 39592832 Free PMC article.
-
mRNA vaccination drives differential mucosal neutralizing antibody profiles in naïve and SARS-CoV-2 previously-infected individuals.Front Immunol. 2022 Sep 8;13:953949. doi: 10.3389/fimmu.2022.953949. eCollection 2022. Front Immunol. 2022. PMID: 36159846 Free PMC article.
-
Reconsideration of Antinucleocapsid IgG Antibody as a Marker of SARS-CoV-2 Infection Postvaccination for Mild COVID-19 Patients.Open Forum Infect Dis. 2022 Dec 15;10(1):ofac677. doi: 10.1093/ofid/ofac677. eCollection 2023 Jan. Open Forum Infect Dis. 2022. PMID: 36655185 Free PMC article.
-
Application of machine learning models to identify serological predictors of COVID-19 severity and outcomes.Res Sq [Preprint]. 2023 Nov 13:rs.3.rs-3463155. doi: 10.21203/rs.3.rs-3463155/v1. Res Sq. 2023. Update in: Commun Med (Lond). 2024 Nov 26;4(1):249. doi: 10.1038/s43856-024-00658-w. PMID: 38014049 Free PMC article. Updated. Preprint.
References
Grants and funding
- U24 OD023382/OD/NIH HHS/United States
- U54 CA260492/CA/NCI NIH HHS/United States
- K23 AI135102/AI/NIAID NIH HHS/United States
- T32 CA009110/CA/NCI NIH HHS/United States
- K08 AI143391/AI/NIAID NIH HHS/United States
- R01 AI130066/AI/NIAID NIH HHS/United States
- R01 NR005228/NR/NINR NIH HHS/United States
- HHSN272201400007C/AI/NIAID NIH HHS/United States
- K08 HS025782/HS/AHRQ HHS/United States
- R21 AI139784/AI/NIAID NIH HHS/United States
- R01 ES026973/ES/NIEHS NIH HHS/United States
- P30 AI094189/AI/NIAID NIH HHS/United States
- U54 EB007958/EB/NIBIB NIH HHS/United States
- UM1 AI068613/AI/NIAID NIH HHS/United States
- U54 HL143541/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

