Quantitative proteomics characterization of cancer biomarkers and treatment
- PMID: 34095463
- PMCID: PMC8142045
- DOI: 10.1016/j.omto.2021.04.006
Quantitative proteomics characterization of cancer biomarkers and treatment
Abstract
Cancer accounted for 16% of all death worldwide in 2018. Significant progress has been made in understanding tumor occurrence, progression, diagnosis, treatment, and prognosis at the molecular level. However, genomics changes cannot truly reflect the state of protein activity in the body due to the poor correlation between genes and proteins. Quantitative proteomics, capable of quantifying the relatively different protein abundance in cancer patients, has been increasingly adopted in cancer research. Quantitative proteomics has great application potentials, including cancer diagnosis, personalized therapeutic drug selection, real-time therapeutic effects and toxicity evaluation, prognosis and drug resistance evaluation, and new therapeutic target discovery. In this review, the development, testing samples, and detection methods of quantitative proteomics are introduced. The biomarkers identified by quantitative proteomics for clinical diagnosis, prognosis, and drug resistance are reviewed. The challenges and prospects of quantitative proteomics for personalized medicine are also discussed.
Keywords: biomarker; cancer; diagnostic marker; quantitative proteomics; therapeutic target.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
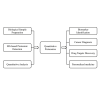
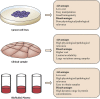
Figures




References
-
- Kristensen V.N., Lingjærde O.C., Russnes H.G., Vollan H.K., Frigessi A., Børresen-Dale A.L. Principles and methods of integrative genomic analyses in cancer. Nat. Rev. Cancer. 2014;14:299–313. - PubMed
-
- Borrebaeck C.A. Precision diagnostics: Moving towards protein biomarker signatures of clinical utility in cancer. Nat. Rev. Cancer. 2017;17:199–204. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

