SARS-CoV-2 detection by a clinical diagnostic RT-LAMP assay
- PMID: 34095506
- PMCID: PMC8170534
- DOI: 10.12688/wellcomeopenres.16517.2
SARS-CoV-2 detection by a clinical diagnostic RT-LAMP assay
Abstract
The ongoing pandemic of SARS-CoV-2 calls for rapid and cost-effective methods to accurately identify infected individuals. The vast majority of patient samples is assessed for viral RNA presence by RT-qPCR. Our biomedical research institute, in collaboration between partner hospitals and an accredited clinical diagnostic laboratory, established a diagnostic testing pipeline that has reported on more than 252,000 RT-qPCR results since its commencement at the beginning of April 2020. However, due to ongoing demand and competition for critical resources, alternative testing strategies were sought. In this work, we present a clinically-validated procedure for high-throughput SARS-CoV-2 detection by RT-LAMP that is robust, reliable, repeatable, specific, and inexpensive.
Keywords: RT-LAMP; SARS-CoV-2; clinical diagnostic.
Copyright: © 2021 Buck MD et al.
Conflict of interest statement
Competing interests: D. Miller and K. Gulati are employees of New England Biolabs, which provided the WarmStart Colorimetric LAMP 2X Master Mix used in this work. C. Swanton receives or has received grant support from Pfizer, AstraZeneca, Bristol-Myers Squibb (BMS), Roche-Ventana, Boehringer-Ingelheim, and Ono Pharmaceutical and has consulted for or received an honorarium from Pfizer, Novartis, GlaxoSmithKline, Merck Sharp & Dohme, BMS, Celgene, AstraZeneca, Illumina, Genentech, Roche-Venatana, GRAIL, Medicxi, and the Sarah Cannon Research Institute. C. Swanton also is a shareholder of Apogen Biotechnologies, Epic Bioscience, and GRAIL and has stock options in and is a cofounder of Achilles Therapeutics.
Figures
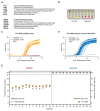
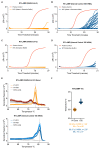
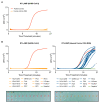

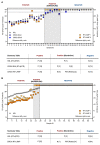
References
-
- Aitken J, Ambrose K, Barrell S, et al. : Scalable and Resilient SARS-CoV-2 testing in an Academic Centre. Health Systems and Quality Improvement. 2020. 10.1101/2020.04.19.20071373 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

