Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch
- PMID: 34095513
- PMCID: PMC8176267
- DOI: 10.12688/wellcomeopenres.16661.2
Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch
Abstract
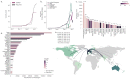
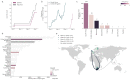
Late in 2020, two genetically-distinct clusters of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with mutations of biological concern were reported, one in the United Kingdom and one in South Africa. Using a combination of data from routine surveillance, genomic sequencing and international travel we track the international dispersal of lineages B.1.1.7 and B.1.351 (variant 501Y-V2). We account for potential biases in genomic surveillance efforts by including passenger volumes from location of where the lineage was first reported, London and South Africa respectively. Using the software tool grinch (global report investigating novel coronavirus haplotypes), we track the international spread of lineages of concern with automated daily reports, Further, we have built a custom tracking website (cov-lineages.org/global_report.html) which hosts this daily report and will continue to include novel SARS-CoV-2 lineages of concern as they are detected.
Keywords: B.1.1.7; B.1.351; N501Y; SARS-CoV-2; air travel; coronavirus; genome sequencing; genomic epidemiology; genomic surveillance; genomics; pandemic; sequencing; surveillance; virus.
Copyright: © 2021 O'Toole Á et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
References
-
- Rambaut A, Loman N, Pybus O, et al. : Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.2020; published online Dec 18. (accessed Jan 8, 2021). Reference Source
-
- Volz E, Mishra S, Chand M, et al. : Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. bioRxiv. 2021. 10.1101/2020.12.30.20249034 - DOI
-
- Tegally H, Wilkinson E, Giovanetti M, et al. : Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. bioRxiv. 2020. 10.1101/2020.12.21.20248640 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

