Redefining Protein Interfaces within Protein Single Crystals with DNA
- PMID: 34096291
- PMCID: PMC8381744
- DOI: 10.1021/jacs.1c04191
Redefining Protein Interfaces within Protein Single Crystals with DNA
Abstract
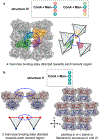
Proteins are exquisite nanoscale building blocks: molecularly pure, chemically addressable, and inherently selective for their evolved function. The organization of proteins into single crystals with high positional, orientational, and translational order results in materials where the location of every atom can be known. However, controlling the organization of proteins is challenging due to the myriad interactions that define protein interfaces within native single crystals. Recently, we discovered that introducing a single DNA-DNA interaction between protein surfaces leads to changes in the packing of proteins within single crystals and the protein-protein interactions (PPIs) that arise. However, modifying specific PPIs to effect deliberate changes to protein packing is an unmet challenge. In this work, we hypothesized that disrupting and replacing a highly conserved PPI with a DNA-DNA interaction would enable protein packing to be modulated by exploiting the programmability of the introduced oligonucleotides. Using concanavalin A (ConA) as a model protein, we circumvent potentially deleterious mutagenesis and exploit the selective binding of ConA toward mannose to noncovalently attach DNA to the protein surface. We show that DNA association eliminates the major PPI responsible for crystallization of native ConA, thereby allowing subtle changes to DNA design (length, complementarity, and attachment position) to program distinct changes to ConA packing, including the realization of three novel crystal structures and the deliberate expansion of ConA packing along a single crystallographic axis. These findings significantly enhance our understanding of how DNA can supersede native PPIs to program protein packing within ordered materials.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Gotti C; Sensini A; Zucchelli A; Carloni R; Focarete ML Hierarchical Fibrous Structures for Muscle-inspired Soft-actuators: A Review. Appl. Mater. Today 2020, 20, 100772.
-
- Bachand GD; Spoerke ED; Stevens MJ Microtubule-Based Nanomaterials: Exploiting Nature’s Dynamic Biopolymers. Biotechnol. Bioeng 2015, 112, 1065–1073. - PubMed
-
- Luo Q; Hou C; Bai Y; Wang R; Liu J Protein Assembly: Versatile Approaches to Construct Highly Ordered Nanostructures. Chem. Rev 2016, 116, 13571–13632. - PubMed

