An essential role of the autophagy activating kinase ULK1 in snRNP biogenesis
- PMID: 34096600
- PMCID: PMC8216288
- DOI: 10.1093/nar/gkab452
An essential role of the autophagy activating kinase ULK1 in snRNP biogenesis
Abstract
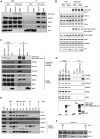
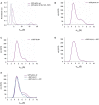
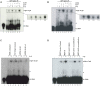
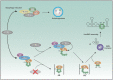
The biogenesis of small uridine-rich nuclear ribonucleoproteins (UsnRNPs) depends on the methylation of Sm proteins catalyzed by the methylosome and the subsequent action of the SMN complex, which assembles the heptameric Sm protein ring onto small nuclear RNAs (snRNAs). In this sophisticated process, the methylosome subunit pICln (chloride conductance regulatory protein) is attributed to an exceptional key position as an 'assembly chaperone' by building up a stable precursor Sm protein ring structure. Here, we show that-apart from its autophagic role-the Ser/Thr kinase ULK1 (Uncoordinated [unc-51] Like Kinase 1) functions as a novel key regulator in UsnRNP biogenesis by phosphorylation of the C-terminus of pICln. As a consequence, phosphorylated pICln is no longer capable to hold up the precursor Sm ring structure. Consequently, inhibition of ULK1 results in a reduction of efficient UsnRNP core assembly. Thus ULK1, depending on its complex formation, exerts different functions in autophagy or snRNP biosynthesis.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Phosphorylation of pICln by the autophagy activating kinase ULK1 regulates snRNP biogenesis and splice activity of the cell.Comput Struct Biotechnol J. 2023 Mar 16;21:2100-2109. doi: 10.1016/j.csbj.2023.03.015. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36968021 Free PMC article.
-
pICln inhibits snRNP biogenesis by binding core spliceosomal proteins.Mol Cell Biol. 1999 Jun;19(6):4113-20. doi: 10.1128/MCB.19.6.4113. Mol Cell Biol. 1999. PMID: 10330151 Free PMC article.
-
Impaired spliceosomal UsnRNP assembly leads to Sm mRNA down-regulation and Sm protein degradation.J Cell Biol. 2017 Aug 7;216(8):2391-2407. doi: 10.1083/jcb.201611108. Epub 2017 Jun 21. J Cell Biol. 2017. PMID: 28637748 Free PMC article.
-
The SMN complex: an assembly machine for RNPs.Cold Spring Harb Symp Quant Biol. 2006;71:313-20. doi: 10.1101/sqb.2006.71.001. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381311 Review.
-
A patent review of UNC-51-like kinase 1/2 inhibitors (2019-present).Expert Opin Ther Pat. 2025 Jan;35(1):7-16. doi: 10.1080/13543776.2024.2423010. Epub 2024 Nov 4. Expert Opin Ther Pat. 2025. PMID: 39470442 Review.
Cited by
-
Development of Macrocyclic PRMT5-Adaptor Protein Interaction Inhibitors.J Med Chem. 2022 Nov 24;65(22):15300-15311. doi: 10.1021/acs.jmedchem.2c01273. Epub 2022 Nov 15. J Med Chem. 2022. PMID: 36378254 Free PMC article.
-
The Impact of p70S6 Kinase-Dependent Phosphorylation of Gemin2 in UsnRNP Biogenesis.Int J Mol Sci. 2023 Oct 25;24(21):15552. doi: 10.3390/ijms242115552. Int J Mol Sci. 2023. PMID: 37958537 Free PMC article.
-
Role of AMPK in autophagy.Front Physiol. 2022 Nov 25;13:1015500. doi: 10.3389/fphys.2022.1015500. eCollection 2022. Front Physiol. 2022. PMID: 36505072 Free PMC article. Review.
-
Phosphorylation of pICln by the autophagy activating kinase ULK1 regulates snRNP biogenesis and splice activity of the cell.Comput Struct Biotechnol J. 2023 Mar 16;21:2100-2109. doi: 10.1016/j.csbj.2023.03.015. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36968021 Free PMC article.
-
The spliceosome pathway activity correlates with reduced anti-tumor immunity and immunotherapy response, and unfavorable clinical outcomes in pan-cancer.Comput Struct Biotechnol J. 2021 Sep 28;19:5428-5442. doi: 10.1016/j.csbj.2021.09.029. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34667536 Free PMC article.
References
-
- Kambach C., Walke S., Young R., Avis J.M., de la Fortelle E., Raker V.A., Luhrmann R., Li J., Nagai K.. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999; 96:375–387. - PubMed
-
- Newman A.J., Nagai K.. Structural studies of the spliceosome: blind men and an elephant. Curr. Opin. Struct. Biol. 2010; 20:82–89. - PubMed
-
- Friesen W.J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G.. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 2002; 277:8243–8247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

