Single-marker and haplotype-based genome-wide association studies for the number of teats in two heavy pig breeds
- PMID: 34096632
- PMCID: PMC8362157
- DOI: 10.1111/age.13095
Single-marker and haplotype-based genome-wide association studies for the number of teats in two heavy pig breeds
Abstract
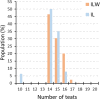
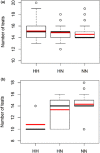
The number of teats is a reproductive-related trait of great economic relevance as it affects the mothering ability of the sows and thus the number of properly weaned piglets. Moreover, genetic improvement of this trait is fundamental to parallelly help the selection for increased litter size. We present the results of single-marker and haplotypes-based genome-wide association studies for the number of teats in two large cohorts of heavy pig breeds (Italian Large White and Italian Landrace) including 3990 animals genotyped with the 70K GGP Porcine BeadChip and other 1927 animals genotyped with the Illumina PorcineSNP60 BeadChip. In the Italian Large White population, genome scans identified three genome regions (SSC7, SSC10, and SSC12) that confirmed the involvement of the VRTN gene (as we previously reported) and highlighted additional loci known to affect teat counts, including the FRMD4A and HOXB1 gene regions. A different picture emerged in the Italian Landrace population, with a total of 12 genome regions in eight chromosomes (SSC3, SSC6, SSC8, SSC11, SSC13, SSC14, SSC15, and SSC16) mainly detected via the haplotype-based genome scan. The most relevant QTL was close to the ARL4C gene on SSC15. Markers in the VRTN gene region were not significant in the Italian Landrace breed. The use of both single-marker and haplotype-based genome-wide association analyses can be helpful to exploit and dissect the genome of the pigs of different populations. Overall, the obtained results supported the polygenic nature of the investigated trait and better elucidated its genetic architecture in Italian heavy pigs.
Keywords: ARL4C; FRMD4A; HOXB1; Sus scrofa; VRTN; Landrace; Large White; morphological trait; single nucleotide polymorphism.
© 2021 The Authors. Animal Genetics published by John Wiley & Sons Ltd on behalf of Stichting International Foundation for Animal Genetics.
Conflict of interest statement
The authors declare they do not have any competing interests.
Figures

Similar articles
-
Genome-wide association studies for the number of teats and teat asymmetry patterns in Large White pigs.Anim Genet. 2020 Aug;51(4):595-600. doi: 10.1111/age.12947. Epub 2020 May 3. Anim Genet. 2020. PMID: 32363597
-
Multiple association studies identify 3 novel candidate genes for teat number trait in Danish Landrace and Large White pigs: BRINP3, LIN52, and UBE3B.J Anim Sci. 2025 Jan 4;103:skaf145. doi: 10.1093/jas/skaf145. J Anim Sci. 2025. PMID: 40305433 Free PMC article.
-
Genome-wide association study of swine farrowing traits. Part II: Bayesian analysis of marker data.J Anim Sci. 2012 Oct;90(10):3360-7. doi: 10.2527/jas.2011-4759. Epub 2012 May 14. J Anim Sci. 2012. PMID: 22585800
-
Genome-wide association study using a single-step approach for teat number in Duroc, Landrace and Yorkshire pigs in Korea.Anim Genet. 2023 Dec;54(6):743-751. doi: 10.1111/age.13357. Epub 2023 Oct 9. Anim Genet. 2023. PMID: 37814452
-
Functional analysis of litter size and number of teats in pigs: From GWAS to post-GWAS.Theriogenology. 2022 Nov;193:157-166. doi: 10.1016/j.theriogenology.2022.09.005. Epub 2022 Sep 19. Theriogenology. 2022. PMID: 36209572
Cited by
-
Genome-wide single nucleotide polymorphism (SNP) data reveal potential candidate genes for litter traits in a Yorkshire pig population.Arch Anim Breed. 2023 Nov 23;66(4):357-368. doi: 10.5194/aab-66-357-2023. eCollection 2023. Arch Anim Breed. 2023. PMID: 38111388 Free PMC article.
-
Genomic Analysis of Reproductive Trait Divergence in Duroc and Yorkshire Pigs: A Comparison of Mixed Models and Selective Sweep Detection.Vet Sci. 2025 Jul 11;12(7):657. doi: 10.3390/vetsci12070657. Vet Sci. 2025. PMID: 40711316 Free PMC article.
-
Genetic diversity and breed-informative SNPs identification in domestic pig populations using coding SNPs.Front Genet. 2023 Nov 16;14:1229741. doi: 10.3389/fgene.2023.1229741. eCollection 2023. Front Genet. 2023. PMID: 38034497 Free PMC article.
-
Genomic-based genetic parameters and genome-wide association studies for productive and reproductive traits in Beef-on-Dairy crossbreds.Front Genet. 2025 Jun 5;16:1530310. doi: 10.3389/fgene.2025.1530310. eCollection 2025. Front Genet. 2025. PMID: 40538636 Free PMC article.
-
Welfare of pigs on farm.EFSA J. 2022 Aug 25;20(8):e07421. doi: 10.2903/j.efsa.2022.7421. eCollection 2022 Aug. EFSA J. 2022. PMID: 36034323 Free PMC article.
References
-
- Arakawa A., Okumura N., Taniguchi M., Hayashi T., Hirose K., Fukawa K., Ito T., Matsumoto T., Uenishi H. & Mikawa S. (2015) Genome‐wide association QTL mapping for teat number in a purebred population of Duroc pigs. Animal Genetics 46, 571–5. - PubMed
-
- Balzani A., Cordell H.J., Sutcliffe E. & Edwards S.A. (2016) Heritability of udder morphology and colostrum quality traits in swine. Journal of Animal Science 94, 3636–44. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

