Cautionary Notes on the Use of Arabinose- and Rhamnose-Inducible Expression Vectors in Pseudomonas aeruginosa
- PMID: 34096777
- PMCID: PMC8297530
- DOI: 10.1128/JB.00224-21
Cautionary Notes on the Use of Arabinose- and Rhamnose-Inducible Expression Vectors in Pseudomonas aeruginosa
Abstract
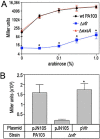
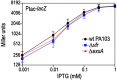
The Pseudomonas aeruginosa virulence factor regulator (Vfr) is a cyclic AMP (cAMP)-responsive transcription factor homologous to the Escherichia coli cAMP receptor protein (CRP). Unlike CRP, which plays a central role in E. coli energy metabolism and catabolite repression, Vfr is primarily involved in the control of P. aeruginosa virulence factor expression. Expression of the Vfr regulon is controlled at the level of vfr transcription, Vfr translation, cAMP synthesis, and cAMP degradation. While investigating mechanisms that regulate Vfr translation, we placed vfr transcription under the control of the rhaBp rhamnose-inducible promoter system (designated PRha) and found that PRha promoter activity was highly dependent upon vfr. Vfr dependence was also observed for the araBp arabinose-inducible promoter (designated PBAD). The observation of Vfr dependence was not entirely unexpected. Both promoters are derived from E. coli, where maximal promoter activity is dependent upon CRP. Like CRP, we found that Vfr directly binds to promoter probes derived from the PRha and PBAD promoters in vitro. Because Vfr-cAMP activity is highly integrated into numerous global regulatory systems, including c-di-GMP signaling, the Gac/Rsm system, MucA/AlgU/AlgZR signaling, and Hfq/sRNAs, the potential exists for significant variability in PRha and PBAD promoter activity in a variety of genetic backgrounds, and use of these promoter systems in P. aeruginosa should be employed with caution. IMPORTANCE Heterologous gene expression and complementation constitute a valuable and widely utilized tool in bacterial genetics. The arabinose-inducible ParaBAD (PBAD) and rhamnose-inducible PrhaBAD (PRha) promoter systems are commonly used in P. aeruginosa genetics and prized for the tight control and dynamic expression ranges that can be achieved. In this study, we demonstrate that the activity of both promoters is dependent upon the cAMP-dependent transcription factor Vfr. While this poses an obvious problem for use in a vfr mutant background, the issue is more pervasive, considering that vfr transcription/synthesis and cAMP homeostasis are highly integrated into the cellular physiology of the organism and influenced by numerous global regulatory systems. Fortunately, the synthetic PTac promoter is not subject to Vfr regulatory control.
Keywords: Pseudomonas aeruginosa; Vfr; arabinose; cyclic AMP; rhamnose.
Figures





Similar articles
-
Characterization of DNA-binding specificity and analysis of binding sites of the Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein.Microbiology (Reading). 2006 Dec;152(Pt 12):3485-3496. doi: 10.1099/mic.0.29008-0. Microbiology (Reading). 2006. PMID: 17159200
-
Hfq and sRNA 179 Inhibit Expression of the Pseudomonas aeruginosa cAMP-Vfr and Type III Secretion Regulons.mBio. 2020 Jun 16;11(3):e00363-20. doi: 10.1128/mBio.00363-20. mBio. 2020. PMID: 32546612 Free PMC article.
-
Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa.Microbiology (Reading). 2002 May;148(Pt 5):1561-1569. doi: 10.1099/00221287-148-5-1561. Microbiology (Reading). 2002. PMID: 11988531
-
Fitting Pieces into the Puzzle of Pseudomonas aeruginosa Type III Secretion System Gene Expression.J Bacteriol. 2019 Jun 10;201(13):e00209-19. doi: 10.1128/JB.00209-19. Print 2019 Jul 1. J Bacteriol. 2019. PMID: 31010903 Free PMC article. Review.
-
Two-component systems required for virulence in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2017 Jun 15;364(11):fnx104. doi: 10.1093/femsle/fnx104. FEMS Microbiol Lett. 2017. PMID: 28510688 Free PMC article. Review.
Cited by
-
Developing a mutant strain of Pseudomonas composti ODT-54 for enhanced production of Psl extracellular polysaccharide.Arch Microbiol. 2025 Mar 7;207(4):81. doi: 10.1007/s00203-025-04292-5. Arch Microbiol. 2025. PMID: 40053128
-
CRISPRi screen identifies FprB as a synergistic target for gallium therapy in Pseudomonas aeruginosa.Nat Commun. 2025 Jul 1;16(1):5870. doi: 10.1038/s41467-025-61208-z. Nat Commun. 2025. PMID: 40595632 Free PMC article.
-
Optimized CRISPR Interference System for Investigating Pseudomonas alloputida Genes Involved in Rhizosphere Microbiome Assembly.ACS Synth Biol. 2024 Sep 20;13(9):2912-2925. doi: 10.1021/acssynbio.4c00312. Epub 2024 Aug 20. ACS Synth Biol. 2024. PMID: 39163848 Free PMC article.
-
Pseudomonas aeruginosa Can Diversify after Host Cell Invasion to Establish Multiple Intracellular Niches.mBio. 2022 Dec 20;13(6):e0274222. doi: 10.1128/mbio.02742-22. Epub 2022 Nov 14. mBio. 2022. PMID: 36374039 Free PMC article.
-
Expansion of healthcare-associated hypervirulent KPC-2-producing Klebsiella pneumoniae ST11/KL64 beyond hospital settings.One Health. 2023 Jun 26;17:100594. doi: 10.1016/j.onehlt.2023.100594. eCollection 2023 Dec. One Health. 2023. PMID: 37448770 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

