Identification and Enzymatic Analysis of an Archaeal ATP-Dependent Serine Kinase from the Hyperthermophilic Archaeon Staphylothermus marinus
- PMID: 34096778
- PMCID: PMC8297531
- DOI: 10.1128/JB.00025-21
Identification and Enzymatic Analysis of an Archaeal ATP-Dependent Serine Kinase from the Hyperthermophilic Archaeon Staphylothermus marinus
Abstract
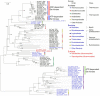
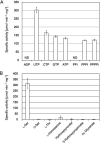
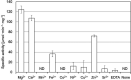
Serine kinase catalyzes the phosphorylation of free serine (Ser) to produce O-phosphoserine (Sep). An ADP-dependent Ser kinase in the hyperthermophilic archaeon Thermococcus kodakarensis (Tk-SerK) is involved in cysteine (Cys) biosynthesis and most likely Ser assimilation. An ATP-dependent Ser kinase in the mesophilic bacterium Staphylococcus aureus is involved in siderophore biosynthesis. Although proteins displaying various degrees of similarity with Tk-SerK are distributed in a wide range of organisms, it is unclear if they are actually Ser kinases. Here, we examined proteins from Desulfurococcales species in Crenarchaeota that display moderate similarity with Tk-SerK from Euryarchaeota (42 to 45% identical). Tk-serK homologs from Staphylothermus marinus (Smar_0555), Desulfurococcus amylolyticus (DKAM_0858), and Desulfurococcus mucosus (Desmu_0904) were expressed in Escherichia coli. All three partially purified recombinant proteins exhibited Ser kinase activity utilizing ATP rather than ADP as a phosphate donor. Purified Smar_0555 protein displayed activity for l-Ser but not other compounds, including d-Ser, l-threonine, and l-homoserine. The enzyme utilized ATP, UTP, GTP, CTP, and the inorganic polyphosphates triphosphate and tetraphosphate as phosphate donors. Kinetic analysis indicated that the Smar_0555 protein preferred nucleoside 5'-triphosphates over triphosphate as a phosphate donor. Transcript levels and Ser kinase activity in S. marinus cells grown with or without serine suggested that the Smar_0555 gene is constitutively expressed. The genes encoding Ser kinases examined here form an operon with genes most likely responsible for the conversion between Sep and 3-phosphoglycerate of central sugar metabolism, suggesting that the ATP-dependent Ser kinases from Desulfurococcales play a role in the assimilation of Ser. IMPORTANCE Homologs of the ADP-dependent Ser kinase from the archaeon Thermococcus kodakarensis (Tk-SerK) include representatives from all three domains of life. The results of this study show that even homologs from the archaeal order Desulfurococcales, which are the most structurally related to the ADP-dependent Ser kinases from the Thermococcales, are Ser kinases that utilize ATP, and in at least some cases inorganic polyphosphates, as the phosphate donor. The differences in properties between the Desulfurococcales and Thermococcales enzymes raise the possibility that Tk-SerK homologs constitute a group of kinases that phosphorylate free serine with a wide range of phosphate donors.
Keywords: Archaea; Crenarchaea; Staphylothermus; enzymes; hyperthermophile; metabolism; serine kinase.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

