Finding Hybrid Incompatibilities Using Genome Sequences from Hybrid Populations
- PMID: 34097068
- PMCID: PMC8476132
- DOI: 10.1093/molbev/msab168
Finding Hybrid Incompatibilities Using Genome Sequences from Hybrid Populations
Abstract
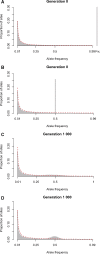
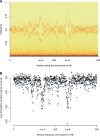
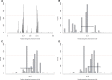
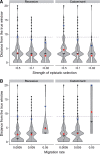
Natural hybrid zones offer a powerful framework for understanding the genetic basis of speciation in progress because ongoing hybridization continually creates unfavorable gene combinations. Evidence indicates that postzygotic reproductive isolation is often caused by epistatic interactions between mutations in different genes that evolved independently of one another (hybrid incompatibilities). We examined the potential to detect epistatic selection against incompatibilities from genome sequence data using the site frequency spectrum (SFS) of polymorphisms by conducting individual-based simulations in SLiM. We found that the genome-wide SFS in hybrid populations assumes a diagnostic shape, with the continual input of fixed differences between source populations via migration inducing a mass at intermediate allele frequency. Epistatic selection locally distorts the SFS as non-incompatibility alleles rise in frequency in a manner analogous to a selective sweep. Building on these results, we present a statistical method to identify genomic regions containing incompatibility loci that locates departures in the local SFS compared with the genome-wide SFS. Cross-validation studies demonstrate that our method detects recessive and codominant incompatibilities across a range of scenarios varying in the strength of epistatic selection, migration rate, and hybrid zone age. Our approach takes advantage of whole genome sequence data, does not require knowledge of demographic history, and can be applied to any pair of nascent species that forms a hybrid zone.
Keywords: epistasis; genetic incompatibilities; hybrid zone; site frequency spectrum.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Barton N, Bengtsson B.. 1986. The barrier to genetic exchange between hybridising populations. Heredity 57(3):357–376. - PubMed
-
- Barton N, Hewitt G.. 1985. Analysis of hybrid zones. Annu Rev Ecol Syst. 16(1):113–148.
-
- Bateson W.1909. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and Modern Science. Cambridge: Cambridge University Press. p. 85–101.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

