Clinical Inflection Point Detection on the Basis of EHR Data to Identify Clinical Trial-Ready Patients With Cancer
- PMID: 34097438
- PMCID: PMC8240790
- DOI: 10.1200/CCI.20.00184
Clinical Inflection Point Detection on the Basis of EHR Data to Identify Clinical Trial-Ready Patients With Cancer
Abstract
Purpose: To inform precision oncology, methods are needed to use electronic health records (EHRs) to identify patients with cancer who are experiencing clinical inflection points, consistent with worsening prognosis or a high propensity to change treatment, at specific time points. Such patients might benefit from real-time screening for clinical trials.
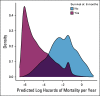
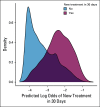
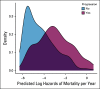
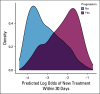
Methods: Using serial unstructured imaging reports for patients with solid tumors or lymphoma participating in a single-institution precision medicine study, we trained a deep neural network natural language processing (NLP) model to dynamically predict patients' prognoses and propensity to start new palliative-intent systemic therapy within 30 days. Model performance was evaluated using Harrell's c-index (for prognosis) and the area under the receiver operating characteristic curve (AUC; for new treatment and new clinical trial enrollment). Associations between model outputs and manual annotations of cancer progression were also evaluated using the AUC.
Results: A deep NLP model was trained and evaluated using 302,688 imaging reports for 16,780 patients. In a held-out test set of 34,770 reports for 1,952 additional patients, the model predicted survival with a c-index of 0.76 and initiation of new treatment with an AUC of 0.77. Model-generated prognostic scores were associated with annotation of cancer progression on the basis of manual EHR review (n = 1,488 reports for 110 patients with lung or colorectal cancer) with an AUC of 0.78, and predictions of new treatment were associated with annotation of cancer progression on the basis of manual EHR review with an AUC of 0.84.
Conclusion: Training a deep NLP model to identify clinical inflection points among patients with cancer is feasible. This approach could identify patients who may benefit from real-time targeted clinical trial screening interventions at health system scale.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

