Hepatitis C Virus Protease Inhibitors Show Differential Efficacy and Interactions with Remdesivir for Treatment of SARS-CoV-2 In Vitro
- PMID: 34097489
- PMCID: PMC8370243
- DOI: 10.1128/AAC.02680-20
Hepatitis C Virus Protease Inhibitors Show Differential Efficacy and Interactions with Remdesivir for Treatment of SARS-CoV-2 In Vitro
Abstract
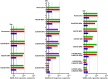
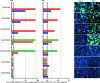
Antivirals targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could improve treatment of COVID-19. We evaluated the efficacy of clinically relevant hepatitis C virus (HCV) NS3 protease inhibitors (PIs) against SARS-CoV-2 and their interactions with remdesivir, the only direct-acting antiviral approved for COVID-19 treatment. HCV PIs showed differential potency in short-term treatment assays based on the detection of SARS-CoV-2 spike protein in Vero E6 cells. Linear PIs boceprevir, telaprevir, and narlaprevir had 50% effective concentrations (EC50) of ∼40 μM. Among the macrocyclic PIs, simeprevir had the highest (EC50, 15 μM) and glecaprevir the lowest (EC50, >178 μM) potency, with paritaprevir, grazoprevir, voxilaprevir, vaniprevir, danoprevir, and deldeprevir in between. Acyclic PIs asunaprevir and faldaprevir had EC50s of 72 and 23 μM, respectively. ACH-806, inhibiting the HCV NS4A protease cofactor, had an EC50 of 46 μM. Similar and slightly increased PI potencies were found in human hepatoma Huh7.5 cells and human lung carcinoma A549-hACE2 cells, respectively. Selectivity indexes based on antiviral and cell viability assays were highest for linear PIs. In short-term treatments, combination of macrocyclic but not linear PIs with remdesivir showed synergism in Vero E6 and A549-hACE2 cells. Longer-term treatment of infected Vero E6 and A549-hACE2 cells with 1-fold EC50 PI revealed minor differences in the barrier to SARS-CoV-2 escape. Viral suppression was achieved with 3- to 8-fold EC50 boceprevir or 1-fold EC50 simeprevir or grazoprevir, but not boceprevir, in combination with 0.4- to 0.8-fold EC50 remdesivir; these concentrations did not lead to viral suppression in single treatments. This study could inform the development and application of protease inhibitors for optimized antiviral treatments of COVID-19.
Keywords: COVID-19; antiviral; combination treatment; coronavirus; repurposing; synergy.
Figures







Similar articles
-
HCV genotype 1-6 NS3 residue 80 substitutions impact protease inhibitor activity and promote viral escape.J Hepatol. 2019 Mar;70(3):388-397. doi: 10.1016/j.jhep.2018.10.031. Epub 2018 Nov 3. J Hepatol. 2019. PMID: 30395912
-
Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture.Cell Rep. 2021 May 18;35(7):109133. doi: 10.1016/j.celrep.2021.109133. Epub 2021 Apr 27. Cell Rep. 2021. PMID: 33984267 Free PMC article.
-
Effects of simeprevir on the replication of SARS-CoV-2 in vitro and in transgenic hACE2 mice.Int J Antimicrob Agents. 2022 Jan;59(1):106499. doi: 10.1016/j.ijantimicag.2021.106499. Epub 2021 Dec 17. Int J Antimicrob Agents. 2022. PMID: 34929295 Free PMC article.
-
A comparative analysis of remdesivir and other repurposed antivirals against SARS-CoV-2.EMBO Mol Med. 2021 Jan 11;13(1):e13105. doi: 10.15252/emmm.202013105. Epub 2020 Nov 3. EMBO Mol Med. 2021. PMID: 33015938 Free PMC article. Review.
-
[Significance of hepatitis C virus baseline polymorphism during the antiviral therapy].Orv Hetil. 2015 May 24;156(21):849-54. doi: 10.1556/650.2015.30180. Orv Hetil. 2015. PMID: 26038992 Review. Hungarian.
Cited by
-
Drug Combinations as a First Line of Defense against Coronaviruses and Other Emerging Viruses.mBio. 2021 Dec 21;12(6):e0334721. doi: 10.1128/mbio.03347-21. Epub 2021 Dec 21. mBio. 2021. PMID: 34933447 Free PMC article. Review.
-
SARS-CoV-2 Production in a Scalable High Cell Density Bioreactor.Vaccines (Basel). 2021 Jun 29;9(7):706. doi: 10.3390/vaccines9070706. Vaccines (Basel). 2021. PMID: 34209694 Free PMC article.
-
Inhibitors of SARS-CoV-2 PLpro.Front Chem. 2022 Apr 26;10:876212. doi: 10.3389/fchem.2022.876212. eCollection 2022. Front Chem. 2022. PMID: 35559224 Free PMC article. Review.
-
Potential inhibitory properties of structurally modified quercetin/isohamnetin glucosides against SARS-CoV-2 Mpro; molecular docking and dynamics simulation strategies.Inform Med Unlocked. 2023;37:101167. doi: 10.1016/j.imu.2023.101167. Epub 2023 Jan 13. Inform Med Unlocked. 2023. PMID: 36686560 Free PMC article.
-
Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system.Sci Adv. 2022 Dec 21;8(51):eadd7197. doi: 10.1126/sciadv.add7197. Epub 2022 Dec 21. Sci Adv. 2022. PMID: 36542720 Free PMC article.
References
-
- Zhou P, Lou Yang X, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang R, Di Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, Liu L, Shan H, Lei C, Hui DSC, Du B, Li L, Zeng G, Yuen KY, Chen R, Tang C, Wang T, Chen P, Xiang J, Li S, Wang JL, Liang Z, Peng Y, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zhong N, China Medical Treatment Expert Group for Covid-19. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. 10.1056/NEJMoa2002032. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Candys Foundation
- Candys Foundation
- Candys Foundation
- Candys Foundation
- Købmand I Odense Johan og Hanne Weimann Født Seedorffs Legat (Købmand i Odense Johann og Hanne Weimann født Seedorffs Legat)
- Ministry of Science, Innovation and Higher Education | Danish Agency for Science and Higher Education (Danish Agency for Science, Technology and Innovation)
- Ministry of Science, Innovation and Higher Education | Danish Agency for Science and Higher Education (Danish Agency for Science, Technology and Innovation)
- Ministry of Science, Innovation and Higher Education | Danish Agency for Science and Higher Education (Danish Agency for Science, Technology and Innovation)
- Novo Nordisk Fonden (NNF)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

