MAB_2355c Confers Macrolide Resistance in Mycobacterium abscessus by Ribosome Protection
- PMID: 34097497
- PMCID: PMC8373217
- DOI: 10.1128/AAC.00330-21
MAB_2355c Confers Macrolide Resistance in Mycobacterium abscessus by Ribosome Protection
Abstract
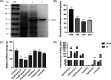
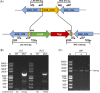
Macrolide resistance is always a concern when treating Mycobacterium abscessus infections. MAB_2355c was identified previously as a possible new factor that confers the intrinsic resistance of 194 clinical M. abscessus isolates to clarithromycin. Herein, the potential mechanism by which MAB_2355c exerts macrolide resistance was explored by bioinformatics analysis, MAB_2355c cloning and protein purification, ATP hydrolysis assay, gene knockout and complementation, antibiotic sensitivity, and transcription-translation assays. MAB_2355c is a putative ATP-binding cassette F (ABC-F) family protein. Purified MAB_2355c protein exhibits ATP hydrolysis activity, which can be inhibited by ribosome-targeting antibiotics. MAB_2355c mRNA expression is upregulated more significantly after exposure to macrolides than after exposure to other ribosome-targeting antibiotics. MAB_2355c deleted strains showed increased sensitivity to macrolides, which was reduced by MAB_2355c complementation. Finally, MAB_2355c rescued the transcription and translation activities affected by macrolides in vitro. These findings suggest that MAB_2355c confers the resistance of M. abscessus to macrolides by ribosome protection, thus complementing other known resistance mechanisms.
Keywords: ABC-F protein; MAB_2355c; Mycobacterium abscessus; macrolides; resistance; ribosome.
Figures



Similar articles
-
Ribosome Protection as a Mechanism of Lincosamide Resistance in Mycobacterium abscessus.Antimicrob Agents Chemother. 2021 Oct 18;65(11):e0118421. doi: 10.1128/AAC.01184-21. Epub 2021 Aug 30. Antimicrob Agents Chemother. 2021. PMID: 34460298 Free PMC article.
-
Mycobacterial HflX is a ribosome splitting factor that mediates antibiotic resistance.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):629-634. doi: 10.1073/pnas.1906748117. Epub 2019 Dec 23. Proc Natl Acad Sci U S A. 2020. PMID: 31871194 Free PMC article.
-
Comparison of drug-susceptibility patterns and gene sequences associated with clarithromycin and azithromycin resistance in Mycobacterium abscessus complex isolates and evaluation of the accumulation of intrinsic macrolide resistance.J Med Microbiol. 2021 Mar;70(3). doi: 10.1099/jmm.0.001326. Epub 2021 Feb 11. J Med Microbiol. 2021. PMID: 33570485
-
Treatment of Mycobacterium abscessus Pulmonary Disease.Chest. 2022 Jan;161(1):64-75. doi: 10.1016/j.chest.2021.07.035. Epub 2021 Jul 24. Chest. 2022. PMID: 34314673 Review.
-
[A pharmacologic approach to treatment of Mycobacterium abscessus pulmonary disease].Rev Mal Respir. 2024 Jan;41(1):29-42. doi: 10.1016/j.rmr.2023.10.010. Epub 2023 Nov 27. Rev Mal Respir. 2024. PMID: 38016833 Review. French.
Cited by
-
The gene MAB_2362 is responsible for intrinsic resistance to various drugs and virulence in Mycobacterium abscessus by regulating cell division.Antimicrob Agents Chemother. 2025 Feb 13;69(2):e0043324. doi: 10.1128/aac.00433-24. Epub 2024 Dec 19. Antimicrob Agents Chemother. 2025. PMID: 39699214 Free PMC article.
-
Exploring antibiotic resistance mechanisms in Mycobacterium abscessus for enhanced therapeutic approaches.Front Microbiol. 2024 Feb 6;15:1331508. doi: 10.3389/fmicb.2024.1331508. eCollection 2024. Front Microbiol. 2024. PMID: 38380095 Free PMC article. Review.
-
Mab2780c, a TetV-like efflux pump, confers high-level spectinomycin resistance in mycobacterium abscessus.Tuberculosis (Edinb). 2023 Jan;138:102295. doi: 10.1016/j.tube.2022.102295. Epub 2022 Dec 22. Tuberculosis (Edinb). 2023. PMID: 36584486 Free PMC article.
-
Lsr2, a pleiotropic regulator at the core of the infectious strategy of Mycobacterium abscessus.Microbiol Spectr. 2024 Mar 5;12(3):e0352823. doi: 10.1128/spectrum.03528-23. Epub 2024 Feb 14. Microbiol Spectr. 2024. PMID: 38353553 Free PMC article.
-
The native ABC-F proteins of Staphylococcus aureus do not contribute to intrinsic resistance against ribosome-targeting antibacterial drugs.J Antimicrob Chemother. 2023 Oct 3;78(10):2601-2603. doi: 10.1093/jac/dkad238. J Antimicrob Chemother. 2023. PMID: 37585343 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

