A human antibody of potent efficacy against SARS-CoV-2 in rhesus macaques showed strong blocking activity to B.1.351
- PMID: 34097570
- PMCID: PMC8189090
- DOI: 10.1080/19420862.2021.1930636
A human antibody of potent efficacy against SARS-CoV-2 in rhesus macaques showed strong blocking activity to B.1.351
Abstract
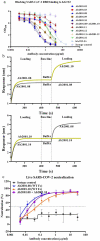
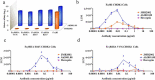
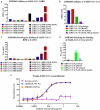
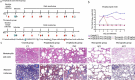
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which causes coronavirus disease-2019 (COVID-19), interacts with the host cell receptor angiotensin-converting enzyme 2 (hACE2) via its spike 1 protein during infection. After the virus sequence was published, we identified two potent antibodies against the SARS-CoV-2 receptor binding domain (RBD) from antibody libraries using a phage-to-yeast (PtY) display platform in only 10 days. Our lead antibody JMB2002, now in a Phase 1 clinical trial (ChiCTR2100042150), showed broad-spectrum in vitro blocking activity against hACE2 binding to the RBD of multiple SARS-CoV-2 variants, including B.1.351 that was reportedly much more resistant to neutralization by convalescent plasma, vaccine sera and some clinical-stage neutralizing antibodies. Furthermore, JMB2002 has demonstrated complete prophylactic and potent therapeutic efficacy in a rhesus macaque disease model. Prophylactic and therapeutic countermeasure intervention of SARS-CoV-2 using JMB2002 would likely slow down the transmission of currently emerged SARS-CoV-2 variants and result in more efficient control of the COVID-19 pandemic.
Keywords: B.1.1.7; B.1.351; D614G; JMB2002; SARS-CoV-2; broad-spectrum; neutralizing antibody; phage-to-yeast; rhesus macaque disease model.
Figures

Similar articles
-
Structural Basis of a Human Neutralizing Antibody Specific to the SARS-CoV-2 Spike Protein Receptor-Binding Domain.Microbiol Spectr. 2021 Oct 31;9(2):e0135221. doi: 10.1128/Spectrum.01352-21. Epub 2021 Oct 13. Microbiol Spectr. 2021. PMID: 34643438 Free PMC article.
-
A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.Nat Commun. 2021 Aug 17;12(1):5000. doi: 10.1038/s41467-021-25331-x. Nat Commun. 2021. PMID: 34404805 Free PMC article.
-
The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody.PLoS One. 2021 Jun 23;16(6):e0253487. doi: 10.1371/journal.pone.0253487. eCollection 2021. PLoS One. 2021. PMID: 34161386 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Severe acute respiratory syndrome-coronavirus-2: Current advances in therapeutic targets and drug development.Rev Med Virol. 2021 May;31(3):e2174. doi: 10.1002/rmv.2174. Epub 2020 Sep 23. Rev Med Virol. 2021. PMID: 32965078 Free PMC article. Review.
Cited by
-
Therapeutic antibodies for COVID-19: is a new age of IgM, IgA and bispecific antibodies coming?MAbs. 2022 Jan-Dec;14(1):2031483. doi: 10.1080/19420862.2022.2031483. MAbs. 2022. PMID: 35220888 Free PMC article. Review.
-
Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.Cell Res. 2022 Jul;32(7):609-620. doi: 10.1038/s41422-022-00672-4. Epub 2022 May 31. Cell Res. 2022. PMID: 35641567 Free PMC article.
-
Antibody-mediated neutralization of SARS-CoV-2.Immunity. 2022 Jun 14;55(6):925-944. doi: 10.1016/j.immuni.2022.05.005. Epub 2022 May 13. Immunity. 2022. PMID: 35623355 Free PMC article. Review.
-
Distinct Conformations of SARS-CoV-2 Omicron Spike Protein and Its Interaction with ACE2 and Antibody.Int J Mol Sci. 2023 Feb 14;24(4):3774. doi: 10.3390/ijms24043774. Int J Mol Sci. 2023. PMID: 36835186 Free PMC article. Review.
-
A broad and potent neutralization epitope in SARS-related coronaviruses.Proc Natl Acad Sci U S A. 2022 Jul 19;119(29):e2205784119. doi: 10.1073/pnas.2205784119. Epub 2022 Jun 29. Proc Natl Acad Sci U S A. 2022. PMID: 35767670 Free PMC article.
References
-
- Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong SX, Silk BJ, Armstrong GL, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage-United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(3):95–99. doi:10.15585/mmwr.mm7003e2. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
