Comparative analysis of three studies measuring fluorescence from engineered bacterial genetic constructs
- PMID: 34097703
- PMCID: PMC8183995
- DOI: 10.1371/journal.pone.0252263
Comparative analysis of three studies measuring fluorescence from engineered bacterial genetic constructs
Abstract
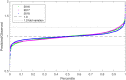
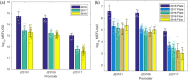
Reproducibility is a key challenge of synthetic biology, but the foundation of reproducibility is only as solid as the reference materials it is built upon. Here we focus on the reproducibility of fluorescence measurements from bacteria transformed with engineered genetic constructs. This comparative analysis comprises three large interlaboratory studies using flow cytometry and plate readers, identical genetic constructs, and compatible unit calibration protocols. Across all three studies, we find similarly high precision in the calibrants used for plate readers. We also find that fluorescence measurements agree closely across the flow cytometry results and two years of plate reader results, with an average standard deviation of 1.52-fold, while the third year of plate reader results are consistently shifted by more than an order of magnitude, with an average shift of 28.9-fold. Analyzing possible sources of error indicates this shift is due to incorrect preparation of the fluorescein calibrant. These findings suggest that measuring fluorescence from engineered constructs is highly reproducible, but also that there is a critical need for access to quality controlled fluorescent calibrants for plate readers.
Conflict of interest statement
The authors of this manuscript have read the journal’s policy and have the following competing interests: The authors received no specific commercial funding for this work. The following authors are employed by for-profit companies: Jacob Beal is employed by Raytheon BBN Technologies; Markus Gershater and Vishal Sanchania are employed by Synthace, and their work on this paper was thus indirectly supported by their salaries. This does not alter the authors’ adherence to PLOS ONE policies on sharing data and materials.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

