Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan
- PMID: 34097716
- PMCID: PMC8238201
- DOI: 10.1371/journal.ppat.1009619
Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan
Abstract
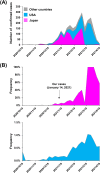
We aimed to investigate novel emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineages in Japan that harbor variants in the spike protein receptor-binding domain (RBD). The total nucleic acid contents of samples from 159 patients with coronavirus disease 2019 (COVID-19) were subjected to whole genome sequencing. The SARS-CoV-2 genome sequences from these patients were examined for variants in spike protein RBD. In January 2021, three family members (one aged in their 40s and two aged under 10 years old) were found to be infected with SARS-CoV-2 harboring W152L/E484K/G769V mutations. These three patients were living in Japan and had no history of traveling abroad. After identifying these cases, we developed a TaqMan assay to screen for the above hallmark mutations and identified an additional 14 patients with the same mutations. The associated virus strain was classified into the GR clade (Global Initiative on Sharing Avian Influenza Data [GISAID]), 20B clade (Nextstrain), and R.1 lineage (Phylogenetic Assignment of Named Global Outbreak [PANGO] Lineages). As of April 22, 2021, R.1 lineage SARS-CoV-2 has been identified in 2,388 SARS-CoV-2 entries in the GISAID database, many of which were from Japan (38.2%; 913/2,388) and the United States (47.1%; 1,125/2,388). Compared with that in the United States, the percentage of SARS-CoV-2 isolates belonging to the R.1 lineage in Japan increased more rapidly over the period from October 24, 2020 to April 18, 2021. R.1 lineage SARS-CoV-2 has potential escape mutations in the spike protein RBD (E484K) and N-terminal domain (W152L); therefore, it will be necessary to continue to monitor the R.1 lineage as it spreads around the world.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
SARS-CoV-2 B.1.1.7 lineage rapidly spreads and replaces R.1 lineage in Japan: Serial and stationary observation in a community.Infect Genet Evol. 2021 Nov;95:105088. doi: 10.1016/j.meegid.2021.105088. Epub 2021 Sep 21. Infect Genet Evol. 2021. PMID: 34560289 Free PMC article.
-
A discernable increase in the severe acute respiratory syndrome coronavirus 2 R.1 lineage carrying an E484K spike protein mutation in Japan.Infect Genet Evol. 2021 Oct;94:105013. doi: 10.1016/j.meegid.2021.105013. Epub 2021 Aug 2. Infect Genet Evol. 2021. PMID: 34352360 Free PMC article.
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
SARS-CoV-2: Evolution and Emergence of New Viral Variants.Viruses. 2022 Mar 22;14(4):653. doi: 10.3390/v14040653. Viruses. 2022. PMID: 35458383 Free PMC article. Review.
-
Recent progress on the mutations of SARS-CoV-2 spike protein and suggestions for prevention and controlling of the pandemic.Infect Genet Evol. 2021 Sep;93:104971. doi: 10.1016/j.meegid.2021.104971. Epub 2021 Jun 17. Infect Genet Evol. 2021. PMID: 34146731 Free PMC article. Review.
Cited by
-
Host Cell Entry and Neutralization Sensitivity of SARS-CoV-2 Lineages B.1.620 and R.1.Viruses. 2022 Nov 9;14(11):2475. doi: 10.3390/v14112475. Viruses. 2022. PMID: 36366573 Free PMC article.
-
SARS-CoV-2 Whole-Genome Sequencing by Ion S5 Technology-Challenges, Protocol Optimization and Success Rates for Different Strains.Viruses. 2022 Jun 6;14(6):1230. doi: 10.3390/v14061230. Viruses. 2022. PMID: 35746701 Free PMC article.
-
SARS-CoV-2 B.1.1.7 lineage rapidly spreads and replaces R.1 lineage in Japan: Serial and stationary observation in a community.Infect Genet Evol. 2021 Nov;95:105088. doi: 10.1016/j.meegid.2021.105088. Epub 2021 Sep 21. Infect Genet Evol. 2021. PMID: 34560289 Free PMC article.
-
The effect of the E484K mutation of SARS-CoV-2 on the neutralizing activity of antibodies from BNT162b2 vaccinated individuals.Vaccine. 2022 Mar 18;40(13):1928-1931. doi: 10.1016/j.vaccine.2022.02.047. Epub 2022 Feb 14. Vaccine. 2022. PMID: 35183387 Free PMC article.
-
Computational modelling of potentially emerging SARS-CoV-2 spike protein RBDs mutations with higher binding affinity towards ACE2: A structural modelling study.Comput Biol Med. 2022 Feb;141:105163. doi: 10.1016/j.compbiomed.2021.105163. Epub 2021 Dec 30. Comput Biol Med. 2022. PMID: 34979405 Free PMC article.
References
-
- Andrew R, Nick L, Oliver P, Wendy B, Jeff B, Alesandro C, et al.. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological 2020. Available from: https://virological.org/t/preliminary-genomic-characterisation-of-an-eme...
-
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al.. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. 10.1101/2020.12.21.20248640 - DOI
-
- Faria NR, Claro IM, Candido D, Franco LAM, Andrade PS, Coletti TM, et al.. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological 2020. Available from: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-c....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

