Genomic analysis of the carboxylesterase family in the salmon louse (Lepeophtheirus salmonis)
- PMID: 34098083
- PMCID: PMC8387733
- DOI: 10.1016/j.cbpc.2021.109095
Genomic analysis of the carboxylesterase family in the salmon louse (Lepeophtheirus salmonis)
Abstract
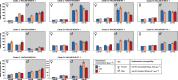
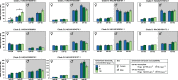
The pyrethroid deltamethrin and the macrocyclic lactone emamectin benzoate (EMB) are used to treat infestations of farmed salmon by parasitic salmon lice, Lepeophtheirus salmonis. While the efficacy of both compounds against Atlantic populations of the parasite has decreased as a result of the evolution of resistance, the molecular mechanisms of drug resistance in L. salmonis are currently not fully understood. The functionally diverse carboxylesterases (CaE) family includes members involved in pesticide resistance phenotypes of terrestrial arthropods. The present study had the objective to characterize the CaE family in L. salmonis and assess its role in drug resistance. L. salmonis CaE homologues were identified by homology searches in the parasite's transcriptome and genome. The transcript expression of CaEs predicted to be catalytically competent was studied using quantitative reverse-transcription PCR in drug susceptible and multi-resistant L. salmonis. The above strategy led to the identification of 21 CaEs genes/pseudogenes. Phylogenetic analyses assigned 13 CaEs to clades involved in neurodevelopmental signaling and cell adhesion, while three sequences were predicted to encode secreted enzymes. Ten CaEs were identified as being potentially catalytically competent. Transcript expression of acetylcholinesterase (ace1b) was significantly increased in multi-resistant lice compared to drug-susceptible L. salmonis, with transcript abundance further increased in preadult-II females following EMB exposure. In summary, results from the present study demonstrate that L. salmonis possesses fewer CaE gene family members than most arthropods characterized so far. Drug resistance in L. salmonis was associated with overexpression of ace1b.
Keywords: Carboxylesterase; Deltamethrin; Emamectin benzoate; Resistance; Salmon lice.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Genome-wide survey of cytochrome P450 genes in the salmon louse Lepeophtheirus salmonis (Krøyer, 1837).Parasit Vectors. 2019 Nov 27;12(1):563. doi: 10.1186/s13071-019-3808-x. Parasit Vectors. 2019. PMID: 31775848 Free PMC article.
-
QTL mapping provides new insights into emamectin benzoate resistance in salmon lice, Lepeophtheirus salmonis.BMC Genomics. 2024 Dec 18;25(1):1212. doi: 10.1186/s12864-024-11096-2. BMC Genomics. 2024. PMID: 39695954 Free PMC article.
-
A 200K SNP chip reveals a novel Pacific salmon louse genotype linked to differential efficacy of emamectin benzoate.Mar Genomics. 2018 Jul;40:45-57. doi: 10.1016/j.margen.2018.03.005. Epub 2018 Apr 16. Mar Genomics. 2018. PMID: 29673959
-
Analysis and management of resistance to chemotherapeutants in salmon lice, Lepeophtheirus salmonis (Copepoda: Caligidae).Pest Manag Sci. 2002 Jun;58(6):528-36. doi: 10.1002/ps.482. Pest Manag Sci. 2002. PMID: 12138619 Review.
-
Avermectin use in aquaculture.Curr Pharm Biotechnol. 2012 May;13(6):1095-102. doi: 10.2174/138920112800399158. Curr Pharm Biotechnol. 2012. PMID: 22039799 Review.
Cited by
-
Key role of mitochondrial mutation Leu107Ser (COX1) in deltamethrin resistance in salmon lice (Lepeophtheirus salmonis).Sci Rep. 2022 Jun 20;12(1):10356. doi: 10.1038/s41598-022-14023-1. Sci Rep. 2022. PMID: 35725748 Free PMC article.
References
-
- Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33:3387–3395. - PubMed
-
- Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. - PubMed
-
- Aranda J., Cerqueira N., Fernandes P.A., Roca M., Tuñón I., Ramos M.J. The catalytic mechanism of carboxylesterases: a computational study. Biochemistry. 2014;53:5820–5829. - PubMed
-
- Auld V.J., Fetter R.D., Broadie K., Goodman C.S. Gliotactin, a novel transmembrane protein on peripheral gila, is required to form the blood-nerve barrier in Drosophila. Cell. 1995;81:757–767. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

