Analysis of the Oxidative Stress Regulon Identifies soxS as a Genetic Target for Resistance Reversal in Multidrug-Resistant Klebsiella pneumoniae
- PMID: 34098732
- PMCID: PMC8262902
- DOI: 10.1128/mBio.00867-21
Analysis of the Oxidative Stress Regulon Identifies soxS as a Genetic Target for Resistance Reversal in Multidrug-Resistant Klebsiella pneumoniae
Abstract
In bacteria, the defense system deployed to counter oxidative stress is orchestrated by three transcriptional factors, SoxS, SoxR, and OxyR. Although the regulon that these factors control is known in many bacteria, similar data are not available for Klebsiella pneumoniae. To address this data gap, oxidative stress was artificially induced in K. pneumoniae MGH78578 using paraquat and the corresponding oxidative stress regulon recorded using transcriptome sequencing (RNA-seq). The soxS gene was significantly induced during oxidative stress, and a knockout mutant was constructed to explore its functionality. The wild type and mutant were grown in the presence of paraquat and subjected to RNA-seq to elucidate the soxS regulon in K. pneumoniae MGH78578. Genes that are commonly regulated both in the oxidative stress and soxS regulons were identified and denoted as the oxidative SoxS regulon; these included a group of genes specifically regulated by SoxS. Efflux pump-encoding genes and global regulators were identified as part of this regulon. Consequently, the isogenic soxS mutant was found to exhibit a reduction in the minimum bactericidal concentration against tetracycline compared to that of the wild type. Impaired efflux activity, allowing tetracycline to be accumulated in the cytoplasm to bactericidal levels, was further evaluated using a tetraphenylphosphonium (TPP+) accumulation assay. The soxS mutant was also susceptible to tetracycline in vivo in a zebrafish embryo model. We conclude that the soxS gene could be considered a genetic target against which an inhibitor could be developed and used in combinatorial therapy to combat infections associated with multidrug-resistant K. pneumoniae. IMPORTANCE Antimicrobial resistance is a global health challenge. Few new antibiotics have been developed for use over the years, and preserving the efficacy of existing compounds is an important step to protect public health. This paper describes a study that examines the effects of exogenously induced oxidative stress on K. pneumoniae and uncovers a target that could be useful to harness as a strategy to mitigate resistance.
Keywords: AMR; Klebsiella pneumoniae; mechanisms of resistance; oxidative stress; soxS.
Figures

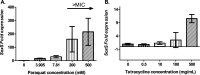
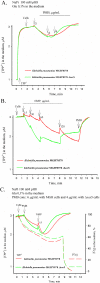

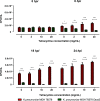
References
-
- Cabiscol E, Tamarit J, Ros J. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

