Identification of COVID-19 B-cell epitopes with phage-displayed peptide library
- PMID: 34098950
- PMCID: PMC8182997
- DOI: 10.1186/s12929-021-00740-8
Identification of COVID-19 B-cell epitopes with phage-displayed peptide library
Abstract
Background: Coronavirus disease 19 (COVID-19) first appeared in the city of Wuhan, in the Hubei province of China. Since its emergence, the COVID-19-causing virus, SARS-CoV-2, has been rapidly transmitted around the globe, overwhelming the medical care systems in many countries and leading to more than 3.3 million deaths. Identification of immunological epitopes on the virus would be highly useful for the development of diagnostic tools and vaccines that will be critical to limiting further spread of COVID-19.
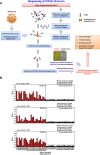
Methods: To find disease-specific B-cell epitopes that correspond to or mimic natural epitopes, we used phage display technology to determine the targets of specific antibodies present in the sera of immune-responsive COVID-19 patients. Enzyme-linked immunosorbent assays were further applied to assess competitive antibody binding and serological detection. VaxiJen, BepiPred-2.0 and DiscoTope 2.0 were utilized for B-cell epitope prediction. PyMOL was used for protein structural analysis.
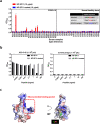
Results: 36 enriched peptides were identified by biopanning with antibodies from two COVID-19 patients; the peptides 4 motifs with consensus residues corresponding to two potential B-cell epitopes on SARS-CoV-2 viral proteins. The putative epitopes and hit peptides were then synthesized for validation by competitive antibody binding and serological detection.
Conclusions: The identified B-cell epitopes on SARS-CoV-2 may aid investigations into COVID-19 pathogenesis and facilitate the development of epitope-based serological diagnostics and vaccines.
Keywords: B-cell epitope; COVID-19; Diagnostic tool; Phage-displayed peptides; SARS-CoV-2; Serological detection.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Center for Systems Science and Engineering (CSSE) at JHU. COVID-19 Dashboard. https://coronavirus.jhu.edu/map.
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

