Conserved Modules Required for Drosophila TRP Function in Vivo
- PMID: 34099505
- PMCID: PMC8265800
- DOI: 10.1523/JNEUROSCI.0200-21.2021
Conserved Modules Required for Drosophila TRP Function in Vivo
Abstract
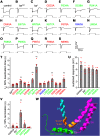
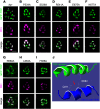
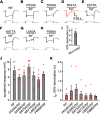
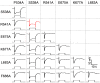
Transient receptor potential (TRP) channels are broadly required in animals for sensory physiology. To provide insights into regulatory mechanisms, the structures of many TRPs have been solved. This has led to new models, some of which have been tested in vitro Here, using the classical TRP required for Drosophila visual transduction, we uncovered structural requirements for channel function in photoreceptor cells. Using a combination of molecular genetics, field recordings, protein expression analysis, and molecular modeling, we interrogated roles for the S4-S5 linker and the TRP domain, and revealed mutations in the S4-S5 linker that impair channel opening or closing. We also uncovered differential requirements for the two highly conserved motifs in the TRP domain for activation and protein stability. By performing genetic complementation, we found an intrasubunit interaction between the S4-S5 linker and the S5 segment that contributes to activation. This analysis highlights key structural requirements for TRP channel opening, closing, folding, and for intrasubunit interactions in a native context-Drosophila photoreceptor cells.SIGNIFICANCE STATEMENT The importance of TRP channels for sensory biology and human health has motivated tremendous effort in trying to understand the roles of the structural motifs essential for their activation, inactivation, and protein folding. In the current work, we have exploited the unique advantages of the Drosophila visual system to reveal mechanistic insights into TRP channel function in a native system-photoreceptor cells. Using a combination of electrophysiology (field recordings), cell biology, and molecular modeling, we have revealed roles of key motifs for activation, inactivation and protein folding of TRP in vivo.
Keywords: Drosophila; TRP channels; TRPC; electroretinogram; phototransduction; vision.
Copyright © 2021 the authors.
Figures