Ribosomal RNA degradation induced by the bacterial RNA polymerase inhibitor rifampicin
- PMID: 34099575
- PMCID: PMC8284325
- DOI: 10.1261/rna.078776.121
Ribosomal RNA degradation induced by the bacterial RNA polymerase inhibitor rifampicin
Abstract
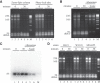
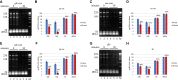
Rifampicin, a broad-spectrum antibiotic, inhibits bacterial RNA polymerase. Here we show that rifampicin treatment of Escherichia coli results in a 50% decrease in cell size due to a terminal cell division. This decrease is a consequence of inhibition of transcription as evidenced by an isogenic rifampicin-resistant strain. There is also a 50% decrease in total RNA due mostly to a 90% decrease in 23S and 16S rRNA levels. Control experiments showed this decrease is not an artifact of our RNA purification protocol and therefore due to degradation in vivo. Since chromosome replication continues after rifampicin treatment, ribonucleotides from rRNA degradation could be recycled for DNA synthesis. Rifampicin-induced rRNA degradation occurs under different growth conditions and in different strain backgrounds. However, rRNA degradation is never complete, thus permitting the reinitiation of growth after removal of rifampicin. The orderly shutdown of growth under conditions where the induction of stress genes is blocked by rifampicin is noteworthy. Inhibition of protein synthesis by chloramphenicol resulted in a partial decrease in 23S and 16S rRNA levels whereas kasugamycin treatment had no effect. Analysis of temperature-sensitive mutant strains implicate RNase E, PNPase, and RNase R in rifampicin-induced rRNA degradation. We cannot distinguish between a direct role for RNase E in rRNA degradation versus an indirect role involving a slowdown of mRNA degradation. Since mRNA and rRNA appear to be degraded by the same ribonucleases, competition by rRNA is likely to result in slower mRNA degradation rates in the presence of rifampicin than under normal growth conditions.
Keywords: PNPase; RNase E; RNase R; cell size; mRNA degradation; ribosomal RNA degradation; rifampicin.
© 2021 Hamouche et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





Similar articles
-
rRNA loss induced by rifampicin addition in Escherichia coli reflects extraction artifacts rather than in vivo degradation.Sci Rep. 2025 Aug 22;15(1):30833. doi: 10.1038/s41598-025-14966-1. Sci Rep. 2025. PMID: 40841809 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Topical antibiotics for chronic suppurative otitis media.Cochrane Database Syst Rev. 2025 Jun 9;6(6):CD013051. doi: 10.1002/14651858.CD013051.pub3. Cochrane Database Syst Rev. 2025. PMID: 40484407
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
Cited by
-
Simulation-based Reconstructed Diffusion unveils the effect of aging on protein diffusion in Escherichia coli.PLoS Comput Biol. 2023 Sep 11;19(9):e1011093. doi: 10.1371/journal.pcbi.1011093. eCollection 2023 Sep. PLoS Comput Biol. 2023. PMID: 37695774 Free PMC article.
-
Challenges and Prospects for Eradication of Helicobacter pylori: Targeting Virulence Factors, Metabolism, and Vaccine Innovation.Pathogens. 2025 Jun 21;14(7):619. doi: 10.3390/pathogens14070619. Pathogens. 2025. PMID: 40732667 Free PMC article. Review.
-
Identification of core biomarkers for tuberculosis progression through bioinformatics analysis and in vitro research.Sci Rep. 2025 Jan 24;15(1):3137. doi: 10.1038/s41598-025-86951-7. Sci Rep. 2025. PMID: 39856190 Free PMC article.
-
Polyribosome-Dependent Clustering of Membrane-Anchored RNA Degradosomes To Form Sites of mRNA Degradation in Escherichia coli.mBio. 2021 Oct 26;12(5):e0193221. doi: 10.1128/mBio.01932-21. Epub 2021 Sep 7. mBio. 2021. PMID: 34488454 Free PMC article.
-
How hydrolytic exoribonucleases impact human disease: Two sides of the same story.FEBS Open Bio. 2023 Jun;13(6):957-974. doi: 10.1002/2211-5463.13392. Epub 2022 Mar 20. FEBS Open Bio. 2023. PMID: 35247037 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
