Leveraging supervised learning for functionally informed fine-mapping of cis-eQTLs identifies an additional 20,913 putative causal eQTLs
- PMID: 34099641
- PMCID: PMC8184741
- DOI: 10.1038/s41467-021-23134-8
Leveraging supervised learning for functionally informed fine-mapping of cis-eQTLs identifies an additional 20,913 putative causal eQTLs
Abstract
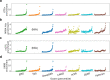
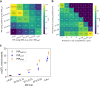
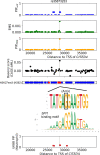
The large majority of variants identified by GWAS are non-coding, motivating detailed characterization of the function of non-coding variants. Experimental methods to assess variants' effect on gene expressions in native chromatin context via direct perturbation are low-throughput. Existing high-throughput computational predictors thus have lacked large gold standard sets of regulatory variants for training and validation. Here, we leverage a set of 14,807 putative causal eQTLs in humans obtained through statistical fine-mapping, and we use 6121 features to directly train a predictor of whether a variant modifies nearby gene expression. We call the resulting prediction the expression modifier score (EMS). We validate EMS by comparing its ability to prioritize functional variants with other major scores. We then use EMS as a prior for statistical fine-mapping of eQTLs to identify an additional 20,913 putatively causal eQTLs, and we incorporate EMS into co-localization analysis to identify 310 additional candidate genes across UK Biobank phenotypes.
Conflict of interest statement
D.G.M. is a founder with equity in Goldfinch Bio, and has received research support from AbbVie, Astellas, Biogen, BioMarin, Eisai, Merck, Pfizer, and Sanofi-Genzyme.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

