B cell signatures and tertiary lymphoid structures contribute to outcome in head and neck squamous cell carcinoma
- PMID: 34099645
- PMCID: PMC8184766
- DOI: 10.1038/s41467-021-23355-x
B cell signatures and tertiary lymphoid structures contribute to outcome in head and neck squamous cell carcinoma
Abstract
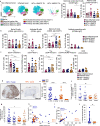
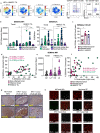
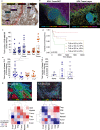
Current immunotherapy paradigms aim to reinvigorate CD8+ T cells, but the contribution of humoral immunity to antitumor immunity remains understudied. Here, we demonstrate that in head and neck squamous cell carcinoma (HNSCC) caused by human papillomavirus infection (HPV+), patients have transcriptional signatures of germinal center (GC) tumor infiltrating B cells (TIL-Bs) and spatial organization of immune cells consistent with tertiary lymphoid structures (TLS) with GCs, both of which correlate with favorable outcome. GC TIL-Bs in HPV+ HNSCC are characterized by distinct waves of gene expression consistent with dark zone, light zone and a transitional state of GC B cells. Semaphorin 4a expression is enhanced on GC TIL-Bs present in TLS of HPV+ HNSCC and during the differentiation of TIL-Bs. Our study suggests that therapeutics to enhance TIL-B responses in HNSCC should be prioritized in future studies to determine if they can complement current T cell mediated immunotherapies.
Conflict of interest statement
D.A.A.V.: cofounder and stockholder—Novasenta, Tizona and Potenza; stockholder—Oncorus and Werewolf; patents licensed and royalties—Astellas, BMS; scientific advisory board member—Tizona, Werewolf and F-Star; consultant—Astellas, BMS, Almirall, Incyte, Bicara; research funding—BMS, Astellas and Novasenta. R.L.F.: a consultant for Aduro Biotech Inc., Bain Capital Life Sciences, Iovance Biotherapeutics Inc., Nanobiotix, Ono Pharmaceutical Co. Ltd., Torque Therapeutics Inc., and TTMS; is on the advisory board for Amgen, AstraZeneca/MedImmune, Bristol-Myers Squibb, EMB Serono, GlaxoSmithKline, Lilly, MacroGenics, Merck, Numab Therapeutics AG, Pfizer, PPD, Regeneron Pharmaceuticals Inc., and Tesaro; receives clinical trial support from AstraZeneca/MedImmune, Bristol-Myers Squibb, and Merck; and receives research funding from Astra-Zeneca/MedImmune, Bristol-Myers Squibb, Tesaro, TTMS, and VentiRx Pharmaceuticals. T.C.B.: Receives research support from Alkermes and Pfizer. A.T.R., A.R.C., T.T., A.L., SaO, S.R.K., C.L., H.A., C.H.L.K., Z.Q., I.A., U.D., S.K., StO, L.G.C., R.S., R.L. declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

